A 2.8-Angstrom-Resolution Cryo-Electron Microscopy Structure of Human Parechovirus 3 in Complex with Fab from a Neutralizing Antibody
- PMID: 30463974
- PMCID: PMC6364030
- DOI: 10.1128/JVI.01597-18
A 2.8-Angstrom-Resolution Cryo-Electron Microscopy Structure of Human Parechovirus 3 in Complex with Fab from a Neutralizing Antibody
Abstract
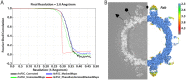
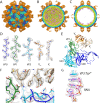
Human parechovirus 3 (HPeV3) infection is associated with sepsis characterized by significant immune activation and subsequent tissue damage in neonates. Strategies to limit infection have been unsuccessful due to inadequate molecular diagnostic tools for early detection and the lack of a vaccine or specific antiviral therapy. Toward the latter, we present a 2.8-Å-resolution structure of HPeV3 in complex with fragments from a neutralizing human monoclonal antibody, AT12-015, using cryo-electron microscopy (cryo-EM) and image reconstruction. Modeling revealed that the epitope extends across neighboring asymmetric units with contributions from capsid proteins VP0, VP1, and VP3. Antibody decoration was found to block binding of HPeV3 to cultured cells. Additionally, at high resolution, it was possible to model a stretch of RNA inside the virion and, from this, identify the key features that drive and stabilize protein-RNA association during assembly.IMPORTANCE Human parechovirus 3 (HPeV3) is receiving increasing attention as a prevalent cause of sepsis-like symptoms in neonates, for which, despite the severity of disease, there are no effective treatments available. Structural and molecular insights into virus neutralization are urgently needed, especially as clinical cases are on the rise. Toward this goal, we present the first structure of HPeV3 in complex with fragments from a neutralizing monoclonal antibody. At high resolution, it was possible to precisely define the epitope that, when targeted, prevents virions from binding to cells. Such an atomic-level description is useful for understanding host-pathogen interactions and viral pathogenesis mechanisms and for finding potential cures for infection and disease.
Keywords: cryo-EM; genome packaging; human parechovirus 3; neutralizing antibodies; picornavirus.
Copyright © 2019 American Society for Microbiology.
Figures


Similar articles
-
Structural Basis of Human Parechovirus Neutralization by Human Monoclonal Antibodies.J Virol. 2015 Sep;89(18):9571-80. doi: 10.1128/JVI.01429-15. Epub 2015 Jul 8. J Virol. 2015. PMID: 26157123 Free PMC article.
-
Near-Atomic Resolution Structure of a Highly Neutralizing Fab Bound to Canine Parvovirus.J Virol. 2016 Oct 14;90(21):9733-9742. doi: 10.1128/JVI.01112-16. Print 2016 Nov 1. J Virol. 2016. PMID: 27535057 Free PMC article.
-
Strain-dependent neutralization reveals antigenic variation of human parechovirus 3.Sci Rep. 2017 Sep 21;7(1):12075. doi: 10.1038/s41598-017-12458-5. Sci Rep. 2017. PMID: 28935894 Free PMC article.
-
Human parechovirus type 3 infection: An emerging infection in neonates and young infants.J Infect Chemother. 2017 Jul;23(7):419-426. doi: 10.1016/j.jiac.2017.04.009. Epub 2017 May 13. J Infect Chemother. 2017. PMID: 28511987 Review.
-
Functional assessment and structural basis of antibody binding to human papillomavirus capsid.Rev Med Virol. 2016 Mar;26(2):115-28. doi: 10.1002/rmv.1867. Epub 2015 Dec 17. Rev Med Virol. 2016. PMID: 26676802 Review.
Cited by
-
Assembly of infectious enteroviruses depends on multiple, conserved genomic RNA-coat protein contacts.PLoS Pathog. 2020 Dec 28;16(12):e1009146. doi: 10.1371/journal.ppat.1009146. eCollection 2020 Dec. PLoS Pathog. 2020. PMID: 33370422 Free PMC article.
-
Parechovirus A Pathogenesis and the Enigma of Genotype A-3.Viruses. 2019 Nov 14;11(11):1062. doi: 10.3390/v11111062. Viruses. 2019. PMID: 31739613 Free PMC article. Review.
-
Deep Learning to Predict Protein Backbone Structure from High-Resolution Cryo-EM Density Maps.Sci Rep. 2020 Mar 9;10(1):4282. doi: 10.1038/s41598-020-60598-y. Sci Rep. 2020. PMID: 32152330 Free PMC article.
-
Development of Monoclonal Antibodies and Antigen-Capture ELISA for Human Parechovirus Type 3.Microorganisms. 2020 Sep 19;8(9):1437. doi: 10.3390/microorganisms8091437. Microorganisms. 2020. PMID: 32961740 Free PMC article.
-
A comparative analysis of parechovirus protein structures with other picornaviruses.Open Biol. 2021 Jul;11(7):210008. doi: 10.1098/rsob.210008. Epub 2021 Jul 28. Open Biol. 2021. PMID: 34315275 Free PMC article.
References
-
- Khatami A, McMullan BJ, Webber M, Stewart P, Francis S, Timmers KJ, Rodas E, Druce J, Mehta B, Sloggett NA, Cumming G, Papadakis G, Kesson AM. 2015. Sepsis-like disease in infants due to human parechovirus type 3 during an outbreak in Australia. Clin Infect Dis 60:228–236. doi:10.1093/cid/ciu784. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

