Efficient Inhibition of Avian and Seasonal Influenza A Viruses by a Virus-Specific Dicer-Substrate Small Interfering RNA Swarm in Human Monocyte-Derived Macrophages and Dendritic Cells
- PMID: 30463970
- PMCID: PMC6364019
- DOI: 10.1128/JVI.01916-18
Efficient Inhibition of Avian and Seasonal Influenza A Viruses by a Virus-Specific Dicer-Substrate Small Interfering RNA Swarm in Human Monocyte-Derived Macrophages and Dendritic Cells
Abstract
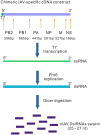
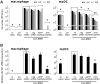
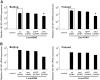
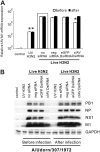
Influenza A viruses (IAVs) are viral pathogens that cause epidemics and occasional pandemics of significant mortality. The generation of efficacious vaccines and antiviral drugs remains a challenge due to the rapid appearance of new influenza virus types and antigenic variants. Consequently, novel strategies for the prevention and treatment of IAV infections are needed, given the limitations of the presently available antivirals. Here, we used enzymatically produced IAV-specific double-stranded RNA (dsRNA) molecules and Giardia intestinalis Dicer for the generation of a swarm of small interfering RNA (siRNA) molecules. The siRNAs target multiple conserved genomic regions of the IAVs. In mammalian cells, the produced 25- to 27-nucleotide-long siRNA molecules are processed by endogenous Dicer into 21-nucleotide siRNAs and are thus designated Dicer-substrate siRNAs (DsiRNAs). We evaluated the efficacy of the above DsiRNA swarm at preventing IAV infections in human primary monocyte-derived macrophages and dendritic cells. The replication of different IAV strains, including avian influenza H5N1 and H7N9 viruses, was significantly inhibited by pretransfection of the cells with the IAV-specific DsiRNA swarm. Up to 7 orders of magnitude inhibition of viral RNA expression was observed, which led to a dramatic inhibition of IAV protein synthesis and virus production. The IAV-specific DsiRNA swarm inhibited virus replication directly through the RNA interference pathway although a weak induction of innate interferon responses was detected. Our results provide direct evidence for the feasibility of the siRNA strategy and the potency of DsiRNA swarms in the prevention and treatment of influenza, including the highly pathogenic avian influenza viruses.IMPORTANCE In spite of the enormous amount of research, influenza virus is still one of the major challenges for medical virology due to its capacity to generate new variants, which potentially lead to severe epidemics and pandemics. We demonstrated here that a swarm of small interfering RNA (siRNA) molecules, including more than 100 different antiviral RNA molecules targeting the most conserved regions of the influenza A virus genome, could efficiently inhibit the replication of all tested avian and seasonal influenza A variants in human primary monocyte-derived macrophages and dendritic cells. The wide antiviral spectrum makes the virus-specific siRNA swarm a potentially efficient treatment modality against both avian and seasonal influenza viruses.
Keywords: Dicer-substrate siRNA; DsiRNA; IAV; IFN; RNA interference; avian influenza virus; gene silencing; human macrophage; human moDC; influenza A virus; interferon response; siRNA swarm; viral replication.
Copyright © 2019 American Society for Microbiology.
Figures

Similar articles
-
Differential Modulation of Innate Immune Responses in Human Primary Cells by Influenza A Viruses Carrying Human or Avian Nonstructural Protein 1.J Virol. 2019 Dec 12;94(1):e00999-19. doi: 10.1128/JVI.00999-19. Print 2019 Dec 12. J Virol. 2019. PMID: 31597767 Free PMC article.
-
Viral Determinants in H5N1 Influenza A Virus Enable Productive Infection of HeLa Cells.J Virol. 2020 Jan 31;94(4):e01410-19. doi: 10.1128/JVI.01410-19. Print 2020 Jan 31. J Virol. 2020. PMID: 31776276 Free PMC article.
-
pH Optimum of Hemagglutinin-Mediated Membrane Fusion Determines Sensitivity of Influenza A Viruses to the Interferon-Induced Antiviral State and IFITMs.J Virol. 2017 May 12;91(11):e00246-17. doi: 10.1128/JVI.00246-17. Print 2017 Jun 1. J Virol. 2017. PMID: 28356532 Free PMC article.
-
In vitro production of synthetic viral RNAs and their delivery into mammalian cells and the application of viral RNAs in the study of innate interferon responses.Methods. 2020 Nov 1;183:21-29. doi: 10.1016/j.ymeth.2019.10.013. Epub 2019 Nov 1. Methods. 2020. PMID: 31682923 Review.
-
Modulation of Innate Immune Responses by the Influenza A NS1 and PA-X Proteins.Viruses. 2018 Dec 12;10(12):708. doi: 10.3390/v10120708. Viruses. 2018. PMID: 30545063 Free PMC article. Review.
Cited by
-
Utilization of Bacteriophage phi6 for the Production of High-Quality Double-Stranded RNA Molecules.Viruses. 2024 Jan 22;16(1):166. doi: 10.3390/v16010166. Viruses. 2024. PMID: 38275976 Free PMC article. Review.
-
Diversity and Current Classification of dsRNA Bacteriophages.Viruses. 2023 Oct 25;15(11):2154. doi: 10.3390/v15112154. Viruses. 2023. PMID: 38005832 Free PMC article. Review.
-
Structural and Functional RNA Motifs of SARS-CoV-2 and Influenza A Virus as a Target of Viral Inhibitors.Int J Mol Sci. 2023 Jan 8;24(2):1232. doi: 10.3390/ijms24021232. Int J Mol Sci. 2023. PMID: 36674746 Free PMC article. Review.
-
Native RNA Purification Method for Small RNA Molecules Based on Asymmetrical Flow Field-Flow Fractionation.Pharmaceuticals (Basel). 2022 Feb 21;15(2):261. doi: 10.3390/ph15020261. Pharmaceuticals (Basel). 2022. PMID: 35215370 Free PMC article.
-
Herpes Simplex Virus Type 1 Clinical Isolates Respond to UL29-Targeted siRNA Swarm Treatment Independent of Their Acyclovir Sensitivity.Viruses. 2020 Dec 13;12(12):1434. doi: 10.3390/v12121434. Viruses. 2020. PMID: 33322225 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

