A Novel Glycoproteomics Workflow Reveals Dynamic O-GlcNAcylation of COPγ1 as a Candidate Regulator of Protein Trafficking
- PMID: 30459710
- PMCID: PMC6232944
- DOI: 10.3389/fendo.2018.00606
A Novel Glycoproteomics Workflow Reveals Dynamic O-GlcNAcylation of COPγ1 as a Candidate Regulator of Protein Trafficking
Abstract
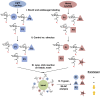
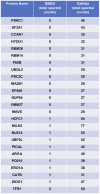
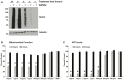
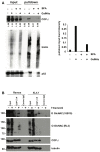
O-linked β-N-acetylglucosamine (O-GlcNAc) is an abundant and essential intracellular form of protein glycosylation in animals and plants. In humans, dysregulation of O-GlcNAcylation occurs in a wide range of diseases, including cancer, diabetes, and neurodegeneration. Since its discovery more than 30 years ago, great strides have been made in understanding central aspects of O-GlcNAc signaling, including identifying thousands of its substrates and characterizing the enzymes that govern it. However, while many O-GlcNAcylated proteins have been reported, only a small subset of these change their glycosylation status in response to a typical stimulus or stress. Identifying the functionally important O-GlcNAcylation changes in any given signaling context remains a significant challenge in the field. To address this need, we leveraged chemical biology and quantitative mass spectrometry methods to create a new glycoproteomics workflow for profiling stimulus-dependent changes in O-GlcNAcylated proteins. In proof-of-principle experiments, we used this new workflow to interrogate changes in O-GlcNAc substrates in mammalian protein trafficking pathways. Interestingly, our results revealed dynamic O-GlcNAcylation of COPγ1, an essential component of the coat protein I (COPI) complex that mediates Golgi protein trafficking. Moreover, we detected 11 O-GlcNAc moieties on COPγ1 and found that this modification is reduced by a model secretory stress that halts COPI trafficking. Our results suggest that O-GlcNAcylation may regulate the mammalian COPI system, analogous to its previously reported roles in other protein trafficking pathways. More broadly, our glycoproteomics workflow is applicable to myriad systems and stimuli, empowering future studies of O-GlcNAc in a host of biological contexts.
Keywords: COPI vesicle trafficking; O-GlcNAc; SILAC; click chemistry; glycoproteomics; protein secretion.
Figures


Similar articles
-
O-GlcNAcylation site mapping by (azide-alkyne) click chemistry and mass spectrometry following intensive fractionation of skeletal muscle cells proteins.J Proteomics. 2018 Aug 30;186:83-97. doi: 10.1016/j.jprot.2018.07.005. Epub 2018 Jul 26. J Proteomics. 2018. PMID: 30016717
-
Evidence for nutrient-dependent regulation of the COPII coat by O-GlcNAcylation.Glycobiology. 2021 Sep 20;31(9):1102-1120. doi: 10.1093/glycob/cwab055. Glycobiology. 2021. PMID: 34142147 Free PMC article.
-
O-GlcNAcylation of amyloid-β precursor protein at threonine 576 residue regulates trafficking and processing.Biochem Biophys Res Commun. 2017 Aug 19;490(2):486-491. doi: 10.1016/j.bbrc.2017.06.067. Epub 2017 Jun 15. Biochem Biophys Res Commun. 2017. PMID: 28624365
-
Emerging roles of O-GlcNAcylation in protein trafficking and secretion.J Biol Chem. 2024 Mar;300(3):105677. doi: 10.1016/j.jbc.2024.105677. Epub 2024 Jan 23. J Biol Chem. 2024. PMID: 38272225 Free PMC article. Review.
-
Detection and identification of O-GlcNAcylated proteins by proteomic approaches.Proteomics. 2015 Mar;15(5-6):1039-50. doi: 10.1002/pmic.201400326. Epub 2015 Feb 3. Proteomics. 2015. PMID: 25429863 Review.
Cited by
-
Directing Traffic: Regulation of COPI Transport by Post-translational Modifications.Front Cell Dev Biol. 2019 Sep 11;7:190. doi: 10.3389/fcell.2019.00190. eCollection 2019. Front Cell Dev Biol. 2019. PMID: 31572722 Free PMC article. Review.
-
Mass Spectrometric Method for the Unambiguous Profiling of Cellular Dynamic Glycosylation.ACS Chem Biol. 2020 Oct 16;15(10):2692-2701. doi: 10.1021/acschembio.0c00453. Epub 2020 Sep 4. ACS Chem Biol. 2020. PMID: 32809798 Free PMC article.
-
Life is sweet: the cell biology of glycoconjugates.Mol Biol Cell. 2019 Mar 1;30(5):525-529. doi: 10.1091/mbc.E18-04-0247. Mol Biol Cell. 2019. PMID: 30817247 Free PMC article. Review.
-
O-GlcNAc Dynamics: The Sweet Side of Protein Trafficking Regulation in Mammalian Cells.Cells. 2023 May 15;12(10):1396. doi: 10.3390/cells12101396. Cells. 2023. PMID: 37408229 Free PMC article. Review.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

