DNA Replication: From Radioisotopes to Click Chemistry
- PMID: 30453631
- PMCID: PMC6278288
- DOI: 10.3390/molecules23113007
DNA Replication: From Radioisotopes to Click Chemistry
Abstract
The replication of nuclear and mitochondrial DNA are basic processes assuring the doubling of the genetic information of eukaryotic cells. In research of the basic principles of DNA replication, and also in the studies focused on the cell cycle, an important role is played by artificially-prepared nucleoside and nucleotide analogues that serve as markers of newly synthesized DNA. These analogues are incorporated into the DNA during DNA replication, and are subsequently visualized. Several methods are used for their detection, including the highly popular click chemistry. This review aims to provide the readers with basic information about the various possibilities of the detection of replication activity using nucleoside and nucleotide analogues, and to show the strengths and weaknesses of those different detection systems, including click chemistry for microscopic studies.
Keywords: click chemistry; indirect immunocytochemistry; isotopes; nucleoside and nucleotide analogues.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures


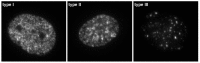
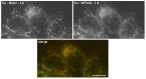

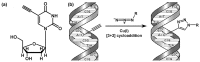
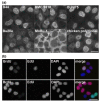

Similar articles
-
Artificial Nucleosides as Diagnostic Probes to Measure Translesion DNA Synthesis.Methods Mol Biol. 2019;1973:237-249. doi: 10.1007/978-1-4939-9216-4_15. Methods Mol Biol. 2019. PMID: 31016706
-
Synthesis of γ-labeled nucleoside 5'-triphosphates using click chemistry.Chem Commun (Camb). 2014 Feb 21;50(15):1861-3. doi: 10.1039/c3cc48937j. Epub 2014 Jan 9. Chem Commun (Camb). 2014. PMID: 24402283
-
Visualizing nucleic acid metabolism using non-natural nucleosides and nucleotide analogs.Biochim Biophys Acta. 2016 Jan;1864(1):165-76. doi: 10.1016/j.bbapap.2015.05.010. Epub 2015 May 22. Biochim Biophys Acta. 2016. PMID: 26004088 Review.
-
Development of a 'clickable' non-natural nucleotide to visualize the replication of non-instructional DNA lesions.Nucleic Acids Res. 2012 Mar;40(5):2357-67. doi: 10.1093/nar/gkr980. Epub 2011 Nov 15. Nucleic Acids Res. 2012. PMID: 22086959 Free PMC article.
-
Click Chemistry and Radiochemistry: The First 10 Years.Bioconjug Chem. 2016 Dec 21;27(12):2791-2807. doi: 10.1021/acs.bioconjchem.6b00561. Epub 2016 Nov 22. Bioconjug Chem. 2016. PMID: 27787983 Free PMC article. Review.
Cited by
-
Basic Methods of Cell Cycle Analysis.Int J Mol Sci. 2023 Feb 12;24(4):3674. doi: 10.3390/ijms24043674. Int J Mol Sci. 2023. PMID: 36835083 Free PMC article. Review.
-
trans-Cyclooctene- and Bicyclononyne-Linked Nucleotides for Click Modification of DNA with Fluorogenic Tetrazines and Live Cell Metabolic Labeling and Imaging.Bioconjug Chem. 2023 Mar 27;34(4):772-80. doi: 10.1021/acs.bioconjchem.3c00064. Online ahead of print. Bioconjug Chem. 2023. PMID: 36972479 Free PMC article.
-
DNA Dyes-Highly Sensitive Reporters of Cell Quantification: Comparison with Other Cell Quantification Methods.Molecules. 2021 Sep 11;26(18):5515. doi: 10.3390/molecules26185515. Molecules. 2021. PMID: 34576986 Free PMC article. Review.
-
Synthetic Thymidine Analog Labeling without Misconceptions.Cells. 2022 Jun 10;11(12):1888. doi: 10.3390/cells11121888. Cells. 2022. PMID: 35741018 Free PMC article.
-
Nucleotide excision repair removes thymidine analog 5-ethynyl-2'-deoxyuridine from the mammalian genome.Proc Natl Acad Sci U S A. 2022 Aug 30;119(35):e2210176119. doi: 10.1073/pnas.2210176119. Epub 2022 Aug 22. Proc Natl Acad Sci U S A. 2022. PMID: 35994676 Free PMC article.
References
-
- Avery O.T., Macleod C.M., McCarty M. Studies on the Chemical Nature of the Substance Inducing Transformation of Pneumococcal Types: Induction of Transformation by a Desoxyribonucleic Acid Fraction Isolated from Pneumococcus Type Iii. J. Exp. Med. 1944;79:137–158. doi: 10.1084/jem.79.2.137. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

