Elevation in Cell Cycle and Protein Metabolism Gene Transcription in Inactive Colonic Tissue From Icelandic Patients With Ulcerative Colitis
- PMID: 30452647
- PMCID: PMC6327231
- DOI: 10.1093/ibd/izy350
Elevation in Cell Cycle and Protein Metabolism Gene Transcription in Inactive Colonic Tissue From Icelandic Patients With Ulcerative Colitis
Abstract
Background: A combination of genetic and environmental factors is thought to be involved in the pathogenesis of ulcerative colitis (UC). In Iceland, the incidence of UC is one of the highest in the world. The aim of this study was to characterize patients with UC and identify potential germline mutations and pathways that could be associated with UC in this population.
Methods: Exome sequencing and genome-wide microarray analysis on macroscopically noninflamed colonic mucosa from patients and controls were performed. Exome sequence data were examined for very rare or novel mutations that were over-represented in the UC cohort. Combined matching of variant analysis and downstream influence on transcriptomic expression in the rectum were analyzed.
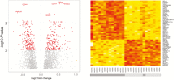
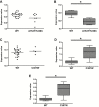
Results: One thousand eight hundred thirty-eight genes were differentially expressed in rectal tissue from UC patients and identified an upregulation in genes associated with cell cycle control and protein processing in the endoplasmic reticulum (ER). Two missense mutations in thiopurine S-methyltransferase (TPMT) with a minor allele frequency of 0.22 in the UC patients compared with a reported 0.062 in the Icelandic population were identified. A predicted damaging mutation in the gene SLC26A3 is potentially associated with increased expression of DUOX2 and DUOXA2 in rectal tissue.
Conclusions: Colonic mucosa of UC patients demonstrates evidence of an elevation in genes involving cell proliferation and processing of proteins within the ER. Exome sequencing identified a possible increased prevalence of 2 damaging TPMT variants within the UC population, suggesting screening the UC population before initiation of thiopurine analogue therapy to avoid toxicity associated with these mutations.
Figures

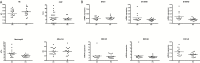
Similar articles
-
Altered endoplasmic reticulum stress affects translation in inactive colon tissue from patients with ulcerative colitis.Gastroenterology. 2011 Sep;141(3):1024-35. doi: 10.1053/j.gastro.2011.05.033. Epub 2011 May 26. Gastroenterology. 2011. PMID: 21699776
-
Colonic Mucosal Transcriptomic Changes in Patients with Long-Duration Ulcerative Colitis Revealed Colitis-Associated Cancer Pathways.J Crohns Colitis. 2019 May 27;13(6):755-763. doi: 10.1093/ecco-jcc/jjz002. J Crohns Colitis. 2019. PMID: 30954025 Free PMC article.
-
Mucosal transcriptomics implicates under expression of BRINP3 in the pathogenesis of ulcerative colitis.Inflamm Bowel Dis. 2014 Oct;20(10):1802-12. doi: 10.1097/MIB.0000000000000169. Inflamm Bowel Dis. 2014. PMID: 25171508 Free PMC article.
-
Transcriptional analysis of the intestinal mucosa of patients with ulcerative colitis in remission reveals lasting epithelial cell alterations.Gut. 2013 Jul;62(7):967-76. doi: 10.1136/gutjnl-2012-303333. Epub 2012 Nov 7. Gut. 2013. PMID: 23135761
-
Difference in Pathomechanism Between Crohn's Disease and Ulcerative Colitis Revealed by Colon Transcriptome.Inflamm Bowel Dis. 2019 Mar 14;25(4):722-731. doi: 10.1093/ibd/izy359. Inflamm Bowel Dis. 2019. PMID: 30517639
Cited by
-
Implication of m6A mRNA Methylation in Susceptibility to Inflammatory Bowel Disease.Epigenomes. 2020 Aug 3;4(3):16. doi: 10.3390/epigenomes4030016. Epigenomes. 2020. PMID: 34968289 Free PMC article.
-
A Novel Role of SLC26A3 in the Maintenance of Intestinal Epithelial Barrier Integrity.Gastroenterology. 2021 Mar;160(4):1240-1255.e3. doi: 10.1053/j.gastro.2020.11.008. Epub 2020 Nov 13. Gastroenterology. 2021. PMID: 33189700 Free PMC article.
-
Molecular Network Analyses Implicate Death-Associated Protein Kinase 3 (DAPK3) as a Key Factor in Colitis-Associated Dysplasia Progression.Inflamm Bowel Dis. 2022 Oct 3;28(10):1485-1496. doi: 10.1093/ibd/izac098. Inflamm Bowel Dis. 2022. PMID: 35604388 Free PMC article.
References
-
- Mirkov MU, Verstockt B, Cleynen I.. Genetics of inflammatory bowel disease: beyond NOD2. Lancet Gastroenterol Hepatol. 2017;2:224–234. - PubMed
-
- Clark PM, Dawany N, Dampier W, et al. . Bioinformatics analysis reveals transcriptome and microrna signatures and drug repositioning targets for IBD and other autoimmune diseases. Inflamm Bowel Dis. 2012;18:2315–2333. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

