Exploring the transcriptome of hormone-naive multifocal prostate cancer and matched lymph node metastases
- PMID: 30449885
- PMCID: PMC6288156
- DOI: 10.1038/s41416-018-0321-5
Exploring the transcriptome of hormone-naive multifocal prostate cancer and matched lymph node metastases
Abstract
Background: The current inability to predict whether a primary prostate cancer (PC) will progress to metastatic disease leads to overtreatment of indolent PCs as well as undertreatment of aggressive PCs. Here, we explored the transcriptional changes associated with metastatic progression of multifocal hormone-naive PC.
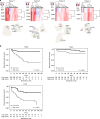
Methods: Using total RNA-sequencing, we analysed laser micro-dissected primary PC foci (n = 23), adjacent normal prostate tissue samples (n = 23) and lymph node metastases (n = 9) from ten hormone-naive PC patients. Genes important for PC progression were identified using differential gene expression and clustering analysis. From these, two multi-gene-based expression signatures (models) were developed, and their prognostic potential was evaluated using Cox-regression and Kaplan-Meier analyses in three independent radical prostatectomy (RP) cohorts (>650 patients).
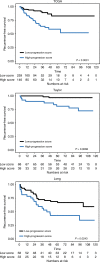
Results: We identified several novel PC-associated transcripts deregulated during PC progression, and these transcripts were used to develop two novel gene-expression-based prognostic models. The models showed independent prognostic potential in three RP cohorts (n = 405, n = 107 and n = 91), using biochemical recurrence after RP as the primary clinical endpoint.
Conclusions: We identified several transcripts deregulated during PC progression and developed two new prognostic models for PC risk stratification, each of which showed independent prognostic value beyond routine clinicopathological factors in three independent RP cohorts.
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Novel diagnostic and prognostic classifiers for prostate cancer identified by genome-wide microRNA profiling.Oncotarget. 2016 May 24;7(21):30760-71. doi: 10.18632/oncotarget.8953. Oncotarget. 2016. PMID: 27120795 Free PMC article.
-
Identifying aggressive prostate cancer foci using a DNA methylation classifier.Genome Biol. 2017 Jan 12;18(1):3. doi: 10.1186/s13059-016-1129-3. Genome Biol. 2017. PMID: 28081708 Free PMC article.
-
DNA methylation signatures for prediction of biochemical recurrence after radical prostatectomy of clinically localized prostate cancer.J Clin Oncol. 2013 Sep 10;31(26):3250-8. doi: 10.1200/JCO.2012.47.1847. Epub 2013 Aug 5. J Clin Oncol. 2013. PMID: 23918943
-
The Role of Prostate-specific Antigen Persistence After Radical Prostatectomy for the Prediction of Clinical Progression and Cancer-specific Mortality in Node-positive Prostate Cancer Patients.Eur Urol. 2016 Jun;69(6):1142-8. doi: 10.1016/j.eururo.2015.12.010. Epub 2015 Dec 31. Eur Urol. 2016. PMID: 26749093
-
Tissue-based Genomics Augments Post-prostatectomy Risk Stratification in a Natural History Cohort of Intermediate- and High-Risk Men.Eur Urol. 2016 Jan;69(1):157-65. doi: 10.1016/j.eururo.2015.05.042. Epub 2015 Jun 6. Eur Urol. 2016. PMID: 26058959
Cited by
-
Stroma-specific gene expression signature identifies prostate cancer subtype with high recurrence risk.NPJ Precis Oncol. 2024 Feb 23;8(1):48. doi: 10.1038/s41698-024-00540-x. NPJ Precis Oncol. 2024. PMID: 38395986 Free PMC article.
-
Genomic Landscape Alterations in Primary Tumor and Matched Lymph Node Metastasis in Hormone-Naïve Prostate Cancer Patients.Cancers (Basel). 2022 Aug 30;14(17):4212. doi: 10.3390/cancers14174212. Cancers (Basel). 2022. PMID: 36077746 Free PMC article.
-
The therapeutic potential of targeting tryptophan catabolism in cancer.Br J Cancer. 2020 Jan;122(1):30-44. doi: 10.1038/s41416-019-0664-6. Epub 2019 Dec 10. Br J Cancer. 2020. PMID: 31819194 Free PMC article. Review.
-
Site-Specific and Common Prostate Cancer Metastasis Genes as Suggested by Meta-Analysis of Gene Expression Data.Life (Basel). 2021 Jun 30;11(7):636. doi: 10.3390/life11070636. Life (Basel). 2021. PMID: 34209195 Free PMC article.
-
Comparative genomics of primary prostate cancer and paired metastases: insights from 12 molecular case studies.J Pathol. 2022 Jul;257(3):274-284. doi: 10.1002/path.5887. Epub 2022 Mar 28. J Pathol. 2022. PMID: 35220606 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

