Identification and characterization of protein N-myristoylation occurring on four human mitochondrial proteins, SAMM50, TOMM40, MIC19, and MIC25
- PMID: 30427857
- PMCID: PMC6235283
- DOI: 10.1371/journal.pone.0206355
Identification and characterization of protein N-myristoylation occurring on four human mitochondrial proteins, SAMM50, TOMM40, MIC19, and MIC25
Abstract
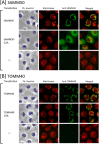
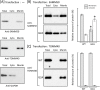
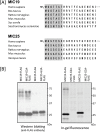
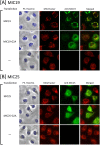
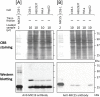
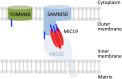
Previously, we showed that SAMM50, a mitochondrial outer membrane protein, is N-myristoylated, and this lipid modification is required for the proper targeting of SAMM50 to mitochondria. In this study, we characterized protein N-myristoylation occurring on four human mitochondrial proteins, SAMM50, TOMM40, MIC19, and MIC25, three of which are components of the mitochondrial intermembrane space bridging (MIB) complex, which plays a critical role in the structure and function of mitochondria. In vitro and in vivo metabolic labeling experiments revealed that all four of these proteins were N-myristoylated. Analysis of intracellular localization of wild-type and non-myristoylated G2A mutants of these proteins by immunofluorescence microscopic analysis and subcellular fractionation analysis indicated that protein N-myristoylation plays a critical role in mitochondrial targeting and membrane binding of two MIB components, SAMM50 and MIC19, but not those of TOMM40 and MIC25. Immunoprecipitation experiments using specific antibodies revealed that MIC19, but not MIC25, was a major N-myristoylated binding partner of SAMM50. Immunoprecipitation experiments using a stable transformant of MIC19 confirmed that protein N-myristoylation of MIC19 is required for the interaction between MIC19 and SAMM50, as reported previously. Thus, protein N-myristoylation occurring on two mitochondrial MIB components, SAMM50 and MIC19, plays a critical role in the mitochondrial targeting and protein-protein interaction between these two MIB components.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures






Similar articles
-
Protein N-myristoylation plays a critical role in the mitochondrial localization of human mitochondrial complex I accessory subunit NDUFB7.Sci Rep. 2023 Dec 27;13(1):22991. doi: 10.1038/s41598-023-50390-z. Sci Rep. 2023. PMID: 38151566 Free PMC article.
-
Identification of Human N-Myristoylated Proteins from Human Complementary DNA Resources by Cell-Free and Cellular Metabolic Labeling Analyses.PLoS One. 2015 Aug 26;10(8):e0136360. doi: 10.1371/journal.pone.0136360. eCollection 2015. PLoS One. 2015. PMID: 26308446 Free PMC article.
-
Sub-mitochondrial localization of the genetic-tagged mitochondrial intermembrane space-bridging components Mic19, Mic60 and Sam50.J Cell Sci. 2017 Oct 1;130(19):3248-3260. doi: 10.1242/jcs.201400. Epub 2017 Aug 14. J Cell Sci. 2017. PMID: 28808085 Free PMC article.
-
Protein myristoylation in protein-lipid and protein-protein interactions.Biophys Chem. 1999 Dec 13;82(2-3):129-37. doi: 10.1016/s0301-4622(99)00112-x. Biophys Chem. 1999. PMID: 10631796 Review.
-
Post-translational myristoylation: Fat matters in cellular life and death.Biochimie. 2011 Jan;93(1):18-31. doi: 10.1016/j.biochi.2010.10.018. Epub 2010 Nov 5. Biochimie. 2011. PMID: 21056615 Review.
Cited by
-
Discovery of Novel Myristic Acid Derivatives as N-Myristoyltransferase Inhibitors: Design, Synthesis, Analysis, Computational Studies and Antifungal Activity.Antibiotics (Basel). 2023 Jul 9;12(7):1167. doi: 10.3390/antibiotics12071167. Antibiotics (Basel). 2023. PMID: 37508263 Free PMC article.
-
Multiomics analysis identifies oxidative phosphorylation as a cancer vulnerability arising from myristoylation inhibition.J Transl Med. 2024 May 7;22(1):431. doi: 10.1186/s12967-024-05150-6. J Transl Med. 2024. PMID: 38715059 Free PMC article.
-
Protein N-myristoylation plays a critical role in the mitochondrial localization of human mitochondrial complex I accessory subunit NDUFB7.Sci Rep. 2023 Dec 27;13(1):22991. doi: 10.1038/s41598-023-50390-z. Sci Rep. 2023. PMID: 38151566 Free PMC article.
-
Discovery of lipid-mediated protein-protein interactions in living cells using metabolic labeling with photoactivatable clickable probes.Chem Sci. 2023 Jan 30;14(9):2419-2430. doi: 10.1039/d2sc06116c. eCollection 2023 Mar 1. Chem Sci. 2023. PMID: 36873846 Free PMC article.
-
A strategy to identify protein-N-myristoylation-dependent phosphorylation reactions of cellular proteins by using Phos-tag SDS-PAGE.PLoS One. 2019 Nov 21;14(11):e0225510. doi: 10.1371/journal.pone.0225510. eCollection 2019. PLoS One. 2019. PMID: 31751425 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

