Contemporary Ribonomics Methods for Viral microRNA Target Analysis
- PMID: 30424002
- PMCID: PMC6316675
- DOI: 10.3390/ncrna4040031
Contemporary Ribonomics Methods for Viral microRNA Target Analysis
Abstract
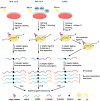
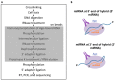
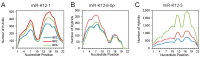
Numerous cellular processes are regulated by microRNAs (miRNAs), both cellular and viral. Elucidating the targets of miRNAs has become an active area of research. An important method in this field is cross-linking and immunoprecipitation (CLIP), where cultured cells or tissues are UV-irradiated to cross-link protein and nucleic acid, the RNA binding protein of interest is immunoprecipitated, and the RNAs pulled down with the protein are isolated, reverse-transcribed, and analyzed by sequencing. CLIP using antibody against Argonaute (Ago), which binds to both miRNA and mRNA as they interact in RISC, has allowed researchers to uncover a large number of miRNA targets. Coupled with high-throughput sequencing, CLIP has been useful for revealing miRNA targetomes for the γ-herpesviruses Kaposi's sarcoma-associated herpesvirus (KSHV) and Epstein-Barr virus (EBV). Variants on the CLIP protocol are described, with the benefits and drawbacks of each. In particular, the most recent methods involving RNA⁻RNA ligation to join the miRNA and its RNA target have aided in target identification. Lastly, data supporting biologically meaningful interactions between miRNAs and long non-coding RNAs (lncRNAs) are reviewed. In summary, ribonomics-based miRNA targetome analysis has expanded our understanding of miRNA targeting and has provided a rich resource for EBV and KSHV research with respect to pathogenesis and tumorigenesis.
Keywords: Ago; CLASH; CLIP; EBV; HITS-CLIP; KSHV; PAR-CLIP; microRNA; qCLASH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Modified Cross-Linking, Ligation, and Sequencing of Hybrids (qCLASH) Identifies Kaposi's Sarcoma-Associated Herpesvirus MicroRNA Targets in Endothelial Cells.J Virol. 2018 Mar 28;92(8):e02138-17. doi: 10.1128/JVI.02138-17. Print 2018 Apr 15. J Virol. 2018. PMID: 29386283 Free PMC article.
-
HITS-CLIP and PAR-CLIP advance viral miRNA targetome analysis.Crit Rev Eukaryot Gene Expr. 2014;24(2):101-16. doi: 10.1615/critreveukaryotgeneexpr.2014006367. Crit Rev Eukaryot Gene Expr. 2014. PMID: 24940765 Free PMC article. Review.
-
Computational analysis of ribonomics datasets identifies long non-coding RNA targets of γ-herpesviral miRNAs.Nucleic Acids Res. 2018 Sep 19;46(16):8574-8589. doi: 10.1093/nar/gky459. Nucleic Acids Res. 2018. PMID: 29846699 Free PMC article.
-
Modified Cross-Linking, Ligation, and Sequencing of Hybrids (qCLASH) to Identify MicroRNA Targets.Curr Protoc. 2021 Oct;1(10):e257. doi: 10.1002/cpz1.257. Curr Protoc. 2021. PMID: 34610213 Free PMC article.
-
Regulation of the MIR155 host gene in physiological and pathological processes.Gene. 2013 Dec 10;532(1):1-12. doi: 10.1016/j.gene.2012.12.009. Epub 2012 Dec 14. Gene. 2013. PMID: 23246696 Review.
Cited by
-
Hybkit: a Python API and command-line toolkit for hybrid sequence data from chimeric RNA methods.Bioinformatics. 2023 Dec 1;39(12):btad721. doi: 10.1093/bioinformatics/btad721. Bioinformatics. 2023. PMID: 38006335 Free PMC article.
-
Cross-Linking Ligation and Sequencing of Hybrids (qCLASH) Reveals an Unpredicted miRNA Targetome in Melanoma Cells.Cancers (Basel). 2021 Mar 4;13(5):1096. doi: 10.3390/cancers13051096. Cancers (Basel). 2021. PMID: 33806450 Free PMC article.
-
MicroRNA Regulation of Human Herpesvirus Latency.Viruses. 2022 Jun 2;14(6):1215. doi: 10.3390/v14061215. Viruses. 2022. PMID: 35746686 Free PMC article. Review.
-
Functional Interplay between RNA Viruses and Non-Coding RNA in Mammals.Noncoding RNA. 2019 Jan 14;5(1):7. doi: 10.3390/ncrna5010007. Noncoding RNA. 2019. PMID: 30646609 Free PMC article. Review.
References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

