Emergence of genotype Cosmopolitan of dengue virus type 2 and genotype III of dengue virus type 3 in Thailand
- PMID: 30419004
- PMCID: PMC6231660
- DOI: 10.1371/journal.pone.0207220
Emergence of genotype Cosmopolitan of dengue virus type 2 and genotype III of dengue virus type 3 in Thailand
Abstract
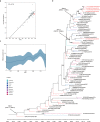
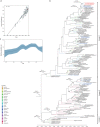
Dengue is a mosquito-borne disease that has spread to over 100 countries. Dengue fever is caused by dengue virus (DENV), which belongs to the Flavivirus genus of the family Flaviviridae. DENV comprises 4 serotypes (DENV-1 to DENV-4), and each serotype is divided into distinct genotypes. Thailand is an endemic area where all 4 serotypes of DENV co-circulate. To understand the current genotype distribution of DENVs in Thailand, we enrolled 100 cases of fever with dengue-like symptoms at the Bamrasnaradura Infectious Diseases Institute during 2016-2017. Among them, 37 cases were shown to be dengue-positive by real-time PCR. We were able to isolate DENVs from 21 cases, including 1 DENV-1, 8 DENV-2, 4 DENV-3, and 8 DENV-4. To investigate the divergence of the viruses, RNA was extracted from isolated DENVs and viral near-whole genome sequences were determined. Phylogenetic analysis of the obtained viral sequences revealed that DENV-2 genotype Cosmopolitan was co-circulating with DENV-2 genotype Asian-I, the previously predominating genotype in Thailand. Furthermore, DENV-3 genotype III was found instead of DENV-3 genotype II. The DENV-2 Cosmopolitan and DENV-3 genotype III found in Thailand were closely related to the respective strains found in nearby countries. These results indicated that DENVs in Thailand have increased in genotypic diversity, and suggested that the DENV genotypic shift observed in other Asian countries also might be taking place in Thailand.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures






Similar articles
-
Unraveling Dengue Virus Diversity in Asia: An Epidemiological Study through Genetic Sequences and Phylogenetic Analysis.Viruses. 2024 Jun 28;16(7):1046. doi: 10.3390/v16071046. Viruses. 2024. PMID: 39066210 Free PMC article. Review.
-
Genotype replacement of dengue virus type 3 and clade replacement of dengue virus type 2 genotype Cosmopolitan in Dhaka, Bangladesh in 2017.Infect Genet Evol. 2019 Nov;75:103977. doi: 10.1016/j.meegid.2019.103977. Epub 2019 Jul 24. Infect Genet Evol. 2019. PMID: 31351235
-
Molecular Characteristics of Dengue Viruses in Patients Hospitalized at the Bamrasnaradura Infectious Diseases Institute, Thailand.Jpn J Infect Dis. 2020 Nov 24;73(6):411-420. doi: 10.7883/yoken.JJID.2020.063. Epub 2020 May 29. Jpn J Infect Dis. 2020. PMID: 32475871
-
Phylogenetic analysis revealed the co-circulation of four dengue virus serotypes in Southern Thailand.PLoS One. 2019 Aug 15;14(8):e0221179. doi: 10.1371/journal.pone.0221179. eCollection 2019. PLoS One. 2019. PMID: 31415663 Free PMC article.
-
Molecular characterization of an imported dengue virus serotype 4 isolate from Thailand.Arch Virol. 2018 Oct;163(10):2903-2906. doi: 10.1007/s00705-018-3906-7. Epub 2018 Jun 14. Arch Virol. 2018. PMID: 29948381 Review.
Cited by
-
Unraveling Dengue Virus Diversity in Asia: An Epidemiological Study through Genetic Sequences and Phylogenetic Analysis.Viruses. 2024 Jun 28;16(7):1046. doi: 10.3390/v16071046. Viruses. 2024. PMID: 39066210 Free PMC article. Review.
-
Molecular surveillance of arboviruses circulation and co-infection during a large chikungunya virus outbreak in Thailand, October 2018 to February 2020.Sci Rep. 2022 Dec 24;12(1):22323. doi: 10.1038/s41598-022-27028-7. Sci Rep. 2022. PMID: 36566236 Free PMC article.
-
Antiviral activities of flavonoids.Biomed Pharmacother. 2021 Aug;140:111596. doi: 10.1016/j.biopha.2021.111596. Epub 2021 Jun 11. Biomed Pharmacother. 2021. PMID: 34126315 Free PMC article. Review.
-
Entomological Surveillance for Zika and Dengue Virus in Aedes Mosquitoes: Implications for Vector Control in Thailand.Pathogens. 2020 Jun 4;9(6):442. doi: 10.3390/pathogens9060442. Pathogens. 2020. PMID: 32512828 Free PMC article.
-
Molecular Characterization of Dengue Virus Strains from the 2019-2020 Epidemic in Hanoi, Vietnam.Microorganisms. 2023 May 11;11(5):1267. doi: 10.3390/microorganisms11051267. Microorganisms. 2023. PMID: 37317240 Free PMC article.
References
-
- Holmes EC, Twiddy SS. The origin, emergence and evolutionary genetics of dengue virus. Infect Genet Evol. 2003;3(1):19–28. - PubMed
-
- Goncalvez AP, Escalante AA, Pujol FH, Ludert JE, Tovar D, Salas RA, et al. Diversity and evolution of the envelope gene of dengue virus type 1. Virology. 2002;303(1):110–9. - PubMed
-
- Twiddy SS, Farrar JJ, Vinh Chau N, Wills B, Gould EA, Gritsun T, et al. Phylogenetic relationships and differential selection pressures among genotypes of dengue-2 virus. Virology. 2002;298(1):63–72. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

