A Nucleosome Bridging Mechanism for Activation of a Maintenance DNA Methyltransferase
- PMID: 30415948
- PMCID: PMC6407616
- DOI: 10.1016/j.molcel.2018.10.006
A Nucleosome Bridging Mechanism for Activation of a Maintenance DNA Methyltransferase
Abstract
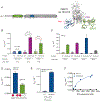
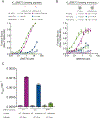
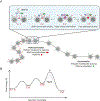
DNA methylation and H3K9me are hallmarks of heterochromatin in plants and mammals, and are successfully maintained across generations. The biochemical and structural basis for this maintenance is poorly understood. The maintenance DNA methyltransferase from Zea mays, ZMET2, recognizes dimethylation of H3K9 via a chromodomain (CD) and a bromo adjacent homology (BAH) domain, which flank the catalytic domain. Here, we show that dinucleosomes are the preferred ZMET2 substrate, with DNA methylation preferentially targeted to linker DNA. Electron microscopy shows one ZMET2 molecule bridging two nucleosomes within a dinucleosome. We find that the CD stabilizes binding, whereas the BAH domain enables allosteric activation by the H3K9me mark. ZMET2 further couples recognition of H3K9me to an increase in the specificity for hemimethylated versus unmethylated DNA. We propose a model in which synergistic coupling between recognition of nucleosome spacing, H3K9 methylation, and DNA modification allows ZMET2 to maintain DNA methylation in heterochromatin with high fidelity.
Keywords: DNA methylation; H3K9me; chromatin; heterochromatin; maize; nucleosome; plants.
Copyright © 2018. Published by Elsevier Inc.
Conflict of interest statement
DECLARATION OF INTERESTS
The authors declare no competing interests.
Figures



Similar articles
-
Dual binding of chromomethylase domains to H3K9me2-containing nucleosomes directs DNA methylation in plants.Cell. 2012 Sep 28;151(1):167-80. doi: 10.1016/j.cell.2012.07.034. Cell. 2012. PMID: 23021223 Free PMC article.
-
Maize chromomethylase Zea methyltransferase2 is required for CpNpG methylation.Plant Cell. 2001 Aug;13(8):1919-28. doi: 10.1105/tpc.010064. Plant Cell. 2001. PMID: 11487702 Free PMC article.
-
Natural variation for alleles under epigenetic control by the maize chromomethylase zmet2.Genetics. 2007 Oct;177(2):749-60. doi: 10.1534/genetics.107.072702. Epub 2007 Jul 29. Genetics. 2007. PMID: 17660570 Free PMC article.
-
Linker histones: novel insights into structure-specific recognition of the nucleosome.Biochem Cell Biol. 2017 Apr;95(2):171-178. doi: 10.1139/bcb-2016-0097. Epub 2016 Jun 29. Biochem Cell Biol. 2017. PMID: 28177778 Free PMC article. Review.
-
Toward an Ensemble View of Chromatosome Structure: A Paradigm Shift from One to Many.Structure. 2018 Aug 7;26(8):1050-1057. doi: 10.1016/j.str.2018.05.009. Epub 2018 Jun 21. Structure. 2018. PMID: 29937356 Review.
Cited by
-
Multi-layered heterochromatin interaction as a switch for DIM2-mediated DNA methylation.Nat Commun. 2024 Aug 9;15(1):6815. doi: 10.1038/s41467-024-51246-4. Nat Commun. 2024. PMID: 39122718 Free PMC article.
-
The unusual predominance of maintenance DNA methylation in Spirodela polyrhiza.G3 (Bethesda). 2024 Apr 3;14(4):jkae004. doi: 10.1093/g3journal/jkae004. G3 (Bethesda). 2024. PMID: 38190722 Free PMC article.
-
Histone H1 prevents non-CG methylation-mediated small RNA biogenesis in Arabidopsis heterochromatin.Elife. 2021 Dec 1;10:e72676. doi: 10.7554/eLife.72676. Elife. 2021. PMID: 34850679 Free PMC article.
-
Detailed insight into the dynamics of the initial phases of de novo RNA-directed DNA methylation in plant cells.Epigenetics Chromatin. 2019 Sep 11;12(1):54. doi: 10.1186/s13072-019-0299-0. Epigenetics Chromatin. 2019. PMID: 31511048 Free PMC article.
-
Depositing centromere repeats induces heritable intragenic heterochromatin establishment and spreading in Arabidopsis.Nucleic Acids Res. 2023 Jul 7;51(12):6039-6054. doi: 10.1093/nar/gkad306. Nucleic Acids Res. 2023. PMID: 37094065 Free PMC article.

