Specificity of the IgG antibody response to Plasmodium falciparum, Plasmodium vivax, Plasmodium malariae, and Plasmodium ovale MSP119 subunit proteins in multiplexed serologic assays
- PMID: 30413163
- PMCID: PMC6230236
- DOI: 10.1186/s12936-018-2566-0
Specificity of the IgG antibody response to Plasmodium falciparum, Plasmodium vivax, Plasmodium malariae, and Plasmodium ovale MSP119 subunit proteins in multiplexed serologic assays
Abstract
Background: Multiplex bead assays (MBA) that measure IgG antibodies to the carboxy-terminal 19-kDa sub-unit of the merozoite surface protein 1 (MSP119) are currently used to determine malaria seroprevalence in human populations living in areas with both stable and unstable transmission. However, the species specificities of the IgG antibody responses to the malaria MSP119 antigens have not been extensively characterized using MBA.
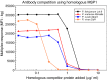
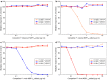
Methods: Recombinant Plasmodium falciparum (3D7), Plasmodium malariae (China I), Plasmodium ovale (Nigeria I), and Plasmodium vivax (Belem) MSP119 proteins were covalently coupled to beads for MBA. Threshold cut-off values for the assays were estimated using sera from US citizens with no history of foreign travel and by receiver operator characteristic curve analysis using diagnostic samples. Banked sera from experimentally infected chimpanzees, sera from humans from low transmission regions of Haiti and Cambodia (N = 12), and elutions from blood spots from humans selected from a high transmission region of Mozambique (N = 20) were used to develop an antigen competition MBA for antibody cross-reactivity studies. A sub-set of samples was further characterized using antibody capture/elution MBA, IgG subclass determination, and antibody avidity measurement.
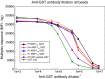
Results: Total IgG antibody responses in experimentally infected chimpanzees were species specific and could be completely suppressed by homologous competitor protein at a concentration of 10 μg/ml. Eleven of 12 samples from the low transmission regions and 12 of 20 samples from the high transmission area had antibody responses that were completely species specific. For 7 additional samples, the P. falciparum MSP119 responses were species specific, but various levels of incomplete heterologous competition were observed for the non-P. falciparum assays. A pan-malaria MSP119 cross-reactive antibody response was observed in elutions of blood spots from two 20-30 years old Mozambique donors. The antibody response from one of these two donors had low avidity and skewed almost entirely to the IgG3 subclass.
Conclusions: Even when P. falciparum, P. malariae, P. ovale, and P. vivax are co-endemic in a high transmission setting, most antibody responses to MSP119 antigens are species-specific and are likely indicative of previous infection history. True pan-malaria cross-reactive responses were found to occur rarely.
Keywords: MSP119; Malaria; Multiplex; Serology; Specificity.
Figures

Similar articles
-
Natural infections with different Plasmodium species induce antibodies reactive to a chimeric Plasmodium vivax recombinant protein.Malar J. 2021 Feb 12;20(1):86. doi: 10.1186/s12936-021-03626-0. Malar J. 2021. PMID: 33579292 Free PMC article.
-
Naturally Acquired Antibody Responses to Plasmodium vivax and Plasmodium falciparum Merozoite Surface Protein 1 (MSP1) C-Terminal 19 kDa Domains in an Area of Unstable Malaria Transmission in Southeast Asia.PLoS One. 2016 Mar 21;11(3):e0151900. doi: 10.1371/journal.pone.0151900. eCollection 2016. PLoS One. 2016. PMID: 26999435 Free PMC article.
-
A panel of recombinant proteins from human-infective Plasmodium species for serological surveillance.Malar J. 2020 Jan 17;19(1):31. doi: 10.1186/s12936-020-3111-5. Malar J. 2020. PMID: 31952523 Free PMC article.
-
Modern immunological approaches to assess malaria transmission and immunity and to diagnose plasmodial infection.Mem Inst Oswaldo Cruz. 1992;87 Suppl 5:117-24. doi: 10.1590/s0074-02761992000900018. Mem Inst Oswaldo Cruz. 1992. PMID: 1342707 Review.
-
Malaria Elimination: Time to Target All Species.Am J Trop Med Hyg. 2018 Jul;99(1):17-23. doi: 10.4269/ajtmh.17-0869. Epub 2018 May 10. Am J Trop Med Hyg. 2018. PMID: 29761762 Free PMC article. Review.
Cited by
-
The use of a chimeric antigen for Plasmodium falciparum and P. vivax seroprevalence estimates from community surveys in Ethiopia and Costa Rica.PLoS One. 2022 May 25;17(5):e0263485. doi: 10.1371/journal.pone.0263485. eCollection 2022. PLoS One. 2022. PMID: 35613090 Free PMC article.
-
Plasmodium vivax malaria serological exposure markers: Assessing the degree and implications of cross-reactivity with P. knowlesi.Cell Rep Med. 2022 Jun 21;3(6):100662. doi: 10.1016/j.xcrm.2022.100662. Cell Rep Med. 2022. PMID: 35732155 Free PMC article.
-
rTgOWP1-f, a specific biomarker for Toxoplasma gondii oocysts.Sci Rep. 2020 May 14;10(1):7947. doi: 10.1038/s41598-020-64590-4. Sci Rep. 2020. PMID: 32409659 Free PMC article.
-
Assessing Performance of HRP2 Antigen Detection for Malaria Diagnosis in Mozambique.J Clin Microbiol. 2019 Aug 26;57(9):e00875-19. doi: 10.1128/JCM.00875-19. Print 2019 Sep. J Clin Microbiol. 2019. PMID: 31270184 Free PMC article.
-
Malaria serology data from the Guiana shield: first insight in IgG antibody responses to Plasmodium falciparum, Plasmodium vivax and Plasmodium malariae antigens in Suriname.Malar J. 2020 Oct 8;19(1):360. doi: 10.1186/s12936-020-03434-y. Malar J. 2020. PMID: 33032606 Free PMC article.
References
-
- Betson M, Clifford S, Stanton M, Kabatereine NB, Stothard JR. Emergence of non falciparum Plasmodium infection despite regular artemisinin combination therapy in an 18-month longitudinal study of Ugandan children and their mothers. J Infect Dis. 2018;217:1099–1109. doi: 10.1093/infdis/jix686. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

