Histone Acetylation Inhibits RSC and Stabilizes the +1 Nucleosome
- PMID: 30401433
- PMCID: PMC6290470
- DOI: 10.1016/j.molcel.2018.09.030
Histone Acetylation Inhibits RSC and Stabilizes the +1 Nucleosome
Abstract
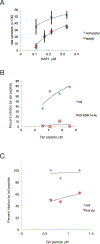
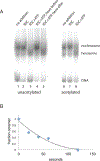
The +1 nucleosome of yeast genes, within which reside transcription start sites, is characterized by histone acetylation, by the displacement of an H2A-H2B dimer, and by a persistent association with the RSC chromatin-remodeling complex. Here we demonstrate the interrelationship of these characteristics and the conversion of a nucleosome to the +1 state in vitro. Contrary to expectation, acetylation performs an inhibitory role, preventing the removal of a nucleosome by RSC. Inhibition is due to both enhanced RSC-histone interaction and diminished histone-chaperone interaction. Acetylation does not prevent all RSC activity, because stably bound RSC removes an H2A-H2B dimer on a timescale of seconds in an irreversible manner.
Keywords: NAP1; NuA4; SAGA; chromatin; chromatin-remodeling; transcription.
Copyright © 2018 Elsevier Inc. All rights reserved.
Conflict of interest statement
DECLARATION OF INTERESTS
The authors declare no competing interests.
Figures


Similar articles
-
Coordinated Action of Nap1 and RSC in Disassembly of Tandem Nucleosomes.Mol Cell Biol. 2016 Aug 12;36(17):2262-71. doi: 10.1128/MCB.00195-16. Print 2016 Sep 1. Mol Cell Biol. 2016. PMID: 27273866 Free PMC article.
-
Histone H3 tail acetylation modulates ATP-dependent remodeling through multiple mechanisms.Nucleic Acids Res. 2011 Oct;39(19):8378-91. doi: 10.1093/nar/gkr535. Epub 2011 Jul 11. Nucleic Acids Res. 2011. PMID: 21749977 Free PMC article.
-
RSC exploits histone acetylation to abrogate the nucleosomal block to RNA polymerase II elongation.Mol Cell. 2006 Nov 3;24(3):481-7. doi: 10.1016/j.molcel.2006.09.012. Mol Cell. 2006. PMID: 17081996 Free PMC article.
-
Spanning the gap: unraveling RSC dynamics in vivo.Curr Genet. 2021 Jun;67(3):399-406. doi: 10.1007/s00294-020-01144-1. Epub 2021 Jan 23. Curr Genet. 2021. PMID: 33484328 Free PMC article. Review.
-
Histone H2B ubiquitination and beyond: Regulation of nucleosome stability, chromatin dynamics and the trans-histone H3 methylation.Epigenetics. 2010 Aug 16;5(6):460-8. doi: 10.4161/epi.5.6.12314. Epub 2010 Aug 16. Epigenetics. 2010. PMID: 20523115 Free PMC article. Review.
Cited by
-
Catching Nucleosome by Its Decorated Tails Determines Its Functional States.Front Genet. 2022 Jul 14;13:903923. doi: 10.3389/fgene.2022.903923. eCollection 2022. Front Genet. 2022. PMID: 35910215 Free PMC article. Review.
-
RSC primes the quiescent genome for hypertranscription upon cell-cycle re-entry.Elife. 2021 May 27;10:e67033. doi: 10.7554/eLife.67033. Elife. 2021. PMID: 34042048 Free PMC article.
-
Interplay among ATP-Dependent Chromatin Remodelers Determines Chromatin Organisation in Yeast.Biology (Basel). 2020 Jul 25;9(8):190. doi: 10.3390/biology9080190. Biology (Basel). 2020. PMID: 32722483 Free PMC article. Review.
-
Poly(dA:dT) Tracts Differentially Modulate Nucleosome Remodeling Activity of RSC and ISW1a Complexes, Exerting Tract Orientation-Dependent and -Independent Effects.Int J Mol Sci. 2023 Oct 17;24(20):15245. doi: 10.3390/ijms242015245. Int J Mol Sci. 2023. PMID: 37894925 Free PMC article.
-
Histone Deacetylase9 Represents the Epigenetic Promotion of M1 Macrophage Polarization and Inflammatory Response via TLR4 Regulation.Biomed Res Int. 2022 Jul 30;2022:7408136. doi: 10.1155/2022/7408136. eCollection 2022. Biomed Res Int. 2022. Retraction in: Biomed Res Int. 2024 Jan 9;2024:9820109. doi: 10.1155/2024/9820109. PMID: 35941971 Free PMC article. Retracted.
References
-
- Belotserkovskaya R, Oh S, Bondarenko VA, Orphanides G, Studitsky VM, and Reinberg D (2003). FACT facilitates transcription-dependent nucleosome alteration. Science 301, 1090–1093. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

