Diverse RNA interference strategies in early-branching metazoans
- PMID: 30382896
- PMCID: PMC6211395
- DOI: 10.1186/s12862-018-1274-2
Diverse RNA interference strategies in early-branching metazoans
Abstract
Background: Micro RNAs (miRNAs) and piwi interacting RNAs (piRNAs), along with the more ancient eukaryotic endogenous small interfering RNAs (endo-siRNAs) constitute the principal components of the RNA interference (RNAi) repertoire of most animals. RNAi in non-bilaterians - sponges, ctenophores, placozoans and cnidarians - appears to be more diverse than that of bilaterians, and includes structurally variable miRNAs in sponges, an enormous number of piRNAs in cnidarians and the absence of miRNAs in ctenophores and placozoans.
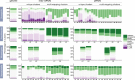
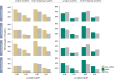
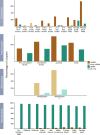
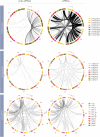
Results: Here we identify thousands of endo-siRNAs and piRNAs from the sponge Amphimedon queenslandica, the ctenophore Mnemiopsis leidyi and the cnidarian Nematostella vectensis using a computational approach that clusters mapped small RNA sequences and annotates each cluster based on the read length and relative abundance of the constituent reads. This approach was validated on 11 small RNA libraries in Drosophila melanogaster, demonstrating the successful annotation of RNAi-associated loci with properties consistent with previous reports. In the non-bilaterians we uncover seven new miRNAs from Amphimedon and four from Nematostella as well as sub-populations of candidate cis-natural antisense transcript (cis-NAT) endo-siRNAs. We confirmed the absence of miRNAs in Mnemiopsis but detected an abundance of endo-siRNAs in this ctenophore. Analysis of putative piRNA structure suggests that conserved localised secondary structures in primary transcripts may be important for the production of mature piRNAs in Amphimedon and Nematostella, as is also the case for endo-siRNAs.
Conclusion: Together, these findings suggest that the last common ancestor of extant animals did not have the entrained RNAi system that typifies bilaterians. Instead it appears that bilaterians, cnidarians, ctenophores and sponges express unique repertoires and combinations of miRNAs, piRNAs and endo-siRNAs.
Keywords: Cnidarian; Ctenophore; Demosponge; Endo-siRNA; Non-bilaterian; RNAi; miRNA; piRNA.
Conflict of interest statement
Ethics approval and consent to participate
No specific ethics approval was required for this project.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Authors' Note A paper was published after acceptance of this manuscript providing evidence for animal-like microRNAs and the miRNA biogenesis machinery in the unicellular ichthyosporeans (Bråte J, Neumann RS, Fromm B, Haraldsen AAB, Tarver, JE., Suga H, et al. (2018). Unicellular Origin of the Animal MicroRNA Machinery. Current Biology, 10.1016/j.cub.2018.08.018). Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
Similar articles
-
The methyltransferase HEN1 is required in Nematostella vectensis for microRNA and piRNA stability as well as larval metamorphosis.PLoS Genet. 2018 Aug 17;14(8):e1007590. doi: 10.1371/journal.pgen.1007590. eCollection 2018 Aug. PLoS Genet. 2018. PMID: 30118479 Free PMC article.
-
Early origins and evolution of microRNAs and Piwi-interacting RNAs in animals.Nature. 2008 Oct 30;455(7217):1193-7. doi: 10.1038/nature07415. Epub 2008 Oct 1. Nature. 2008. PMID: 18830242 Free PMC article.
-
Surprisingly rich repertoire of Wnt genes in the demosponge Halisarca dujardini.BMC Evol Biol. 2016 Jun 10;16(1):123. doi: 10.1186/s12862-016-0700-6. BMC Evol Biol. 2016. PMID: 27287511 Free PMC article.
-
A non-bilaterian perspective on the development and evolution of animal digestive systems.Cell Tissue Res. 2019 Sep;377(3):321-339. doi: 10.1007/s00441-019-03075-x. Epub 2019 Aug 7. Cell Tissue Res. 2019. PMID: 31388768 Free PMC article. Review.
-
Technology of RNA Interference in Advanced Medicine.Microrna. 2018;7(2):74-84. doi: 10.2174/2211536607666180129153307. Microrna. 2018. PMID: 29380708 Review.
Cited by
-
Viral Infection and Stress Affect Protein Levels of Dicer 2 and Argonaute 2 in Drosophila melanogaster.Front Immunol. 2020 Mar 4;11:362. doi: 10.3389/fimmu.2020.00362. eCollection 2020. Front Immunol. 2020. PMID: 32194567 Free PMC article.
-
Exploiting somatic piRNAs in Bemisia tabaci enables novel gene silencing through RNA feeding.Life Sci Alliance. 2020 Aug 6;3(10):e202000731. doi: 10.26508/lsa.202000731. Print 2020 Oct. Life Sci Alliance. 2020. PMID: 32764103 Free PMC article.
-
Unravelling the developmental and functional significance of an ancient Argonaute duplication.Nat Commun. 2020 Dec 3;11(1):6187. doi: 10.1038/s41467-020-20003-8. Nat Commun. 2020. PMID: 33273471 Free PMC article.
-
The origin of RNA interference: Adaptive or neutral evolution?PLoS Biol. 2022 Jun 29;20(6):e3001715. doi: 10.1371/journal.pbio.3001715. eCollection 2022 Jun. PLoS Biol. 2022. PMID: 35767561 Free PMC article.
-
Nuclear Argonaute Piwi Gene Mutation Affects rRNA by Inducing rRNA Fragment Accumulation, Antisense Expression, and Defective Processing in Drosophila Ovaries.Int J Mol Sci. 2020 Feb 7;21(3):1119. doi: 10.3390/ijms21031119. Int J Mol Sci. 2020. PMID: 32046213 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

