Conformational flexibility of histone variant CENP-ACse4 is regulated by histone H4: A mechanism to stabilize soluble Cse4
- PMID: 30381395
- PMCID: PMC6311523
- DOI: 10.1074/jbc.RA118.004141
Conformational flexibility of histone variant CENP-ACse4 is regulated by histone H4: A mechanism to stabilize soluble Cse4
Abstract
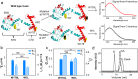
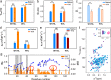
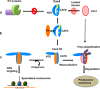
The histone variant CENP-ACse4 is a core component of the specialized nucleosome at the centromere in budding yeast and is required for genomic integrity. Accordingly, the levels of Cse4 in cells are tightly regulated, primarily by ubiquitin-mediated proteolysis. However, structural transitions in Cse4 that regulate its centromeric localization and interaction with regulatory components are poorly understood. Using time-resolved fluorescence, NMR, and molecular dynamics simulations, we show here that soluble Cse4 can exist in a "closed" conformation, inaccessible to various regulatory components. We further determined that binding of its obligate partner, histone H4, alters the interdomain interaction within Cse4, enabling an "open" state that is susceptible to proteolysis. This dynamic model allows kinetochore formation only in the presence of H4, as the Cse4 N terminus, which is required for interaction with other centromeric components, is unavailable in the absence of H4. The specific requirement of H4 binding for the conformational regulation of Cse4 suggests a structure-based regulatory mechanism for Cse4 localization. Our data suggested a novel structural transition-based mechanism where conformational flexibility of the Cse4 N terminus can control Cse4 levels in the yeast cell and prevent Cse4 from interacting with kinetochore components at ectopic locations for formation of premature kinetochore assembly.
Keywords: cell division; centromere; centromeric protein A; chromosome; fluorescence anisotropy; histone; histone tails; histone variants; kinetochore assembly; molecular dynamics; protein-protein interactions; structural biology.
© 2018 Malik et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures


Similar articles
-
Histone H4 Facilitates the Proteolysis of the Budding Yeast CENP-ACse4 Centromeric Histone Variant.Genetics. 2017 Jan;205(1):113-124. doi: 10.1534/genetics.116.194027. Epub 2016 Oct 28. Genetics. 2017. PMID: 27794026 Free PMC article.
-
Structural basis for recognition of centromere histone variant CenH3 by the chaperone Scm3.Nature. 2011 Apr 14;472(7342):234-7. doi: 10.1038/nature09854. Epub 2011 Mar 16. Nature. 2011. PMID: 21412236 Free PMC article.
-
Molecular basis for the selective recognition and ubiquitination of centromeric histone H3 by yeast E3 ligase Psh1.J Genet Genomics. 2021 Jun 20;48(6):463-472. doi: 10.1016/j.jgg.2021.04.007. Epub 2021 May 26. J Genet Genomics. 2021. PMID: 34217622
-
Protein kinases in mitotic phosphorylation of budding yeast CENP-A.Curr Genet. 2019 Dec;65(6):1325-1332. doi: 10.1007/s00294-019-00997-5. Epub 2019 May 22. Curr Genet. 2019. PMID: 31119371 Review.
-
Insights into assembly and regulation of centromeric chromatin in Saccharomyces cerevisiae.Biochim Biophys Acta. 2012 Jul;1819(7):776-83. doi: 10.1016/j.bbagrm.2012.02.008. Epub 2012 Feb 16. Biochim Biophys Acta. 2012. PMID: 22366340 Free PMC article. Review.
Cited by
-
The regulation of chromosome segregation via centromere loops.Crit Rev Biochem Mol Biol. 2019 Aug;54(4):352-370. doi: 10.1080/10409238.2019.1670130. Epub 2019 Oct 1. Crit Rev Biochem Mol Biol. 2019. PMID: 31573359 Free PMC article. Review.
-
Interaction of histone H4 with Cse4 facilitates conformational changes in Cse4 for its sumoylation and mislocalization.Nucleic Acids Res. 2024 Jan 25;52(2):643-659. doi: 10.1093/nar/gkad1133. Nucleic Acids Res. 2024. PMID: 38038247 Free PMC article.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

