DSMNC: a database of somatic mutations in normal cells
- PMID: 30380071
- PMCID: PMC6323907
- DOI: 10.1093/nar/gky1045
DSMNC: a database of somatic mutations in normal cells
Abstract
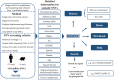
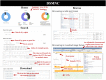
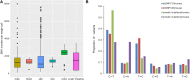
Numerous non-inherited somatic mutations, distinct from those of germ-line origin, occur in somatic cells during DNA replication per cell-division. The somatic mutations, recording the unique genetic cell-lineage 'history' of each proliferating normal cell, are important but remain to be investigated because of their ultra-low frequency hidden in the genetic background of heterogeneous cells. Luckily, the recent development of single-cell genomics biotechnologies enables the screening and collection of the somatic mutations, especial single nucleotide variations (SNVs), occurring in normal cells. Here, we established DSMNC: a database of somatic mutations in normal cells (http://dsmnc.big.ac.cn/), which provides most comprehensive catalogue of somatic SNVs in single cells from various normal tissues. In the current version, the database collected ∼0.8 million SNVs accumulated in ∼600 single normal cells (579 human cells and 39 mouse cells). The database interface supports the user-friendly capability of browsing and searching the SNVs and their annotation information. DSMNC, which serves as a timely and valuable collection of somatic mutations in individual normal cells, has made it possible to analyze the burdens and signatures of somatic mutations in various types of heterogeneous normal cells. Therefore, DSMNC will significantly improve our understanding of the characteristics of somatic mutations in normal cells.
Figures
Similar articles
-
SomaMutDB: a database of somatic mutations in normal human tissues.Nucleic Acids Res. 2022 Jan 7;50(D1):D1100-D1108. doi: 10.1093/nar/gkab914. Nucleic Acids Res. 2022. PMID: 34634815 Free PMC article.
-
Comprehensive identification of somatic nucleotide variants in human brain tissue.Genome Biol. 2021 Mar 29;22(1):92. doi: 10.1186/s13059-021-02285-3. Genome Biol. 2021. PMID: 33781308 Free PMC article.
-
FaSD-somatic: a fast and accurate somatic SNV detection algorithm for cancer genome sequencing data.Bioinformatics. 2014 Sep 1;30(17):2498-500. doi: 10.1093/bioinformatics/btu338. Epub 2014 May 14. Bioinformatics. 2014. PMID: 24833803
-
Mutational signatures: the patterns of somatic mutations hidden in cancer genomes.Curr Opin Genet Dev. 2014 Feb;24(100):52-60. doi: 10.1016/j.gde.2013.11.014. Epub 2013 Dec 29. Curr Opin Genet Dev. 2014. PMID: 24657537 Free PMC article. Review.
-
Direct mutation analysis by high-throughput sequencing: from germline to low-abundant, somatic variants.Mutat Res. 2012 Jan 3;729(1-2):1-15. doi: 10.1016/j.mrfmmm.2011.10.001. Epub 2011 Oct 12. Mutat Res. 2012. PMID: 22016070 Free PMC article. Review.
Cited by
-
Core promoter in TNBC is highly mutated with rich ethnic signature.Brief Funct Genomics. 2023 Jan 20;22(1):9-19. doi: 10.1093/bfgp/elac035. Brief Funct Genomics. 2023. PMID: 36307127 Free PMC article.
-
Different Oncogenes and Reproductive Histories Shape the Progression of Distinct Premalignant Clones in Multistage Mouse Breast Cancer Models.Am J Pathol. 2024 Jul;194(7):1329-1345. doi: 10.1016/j.ajpath.2024.02.018. Epub 2024 Mar 25. Am J Pathol. 2024. PMID: 38537934
-
Pan-cancer analyses of synonymous mutations based on tissue-specific codon optimality.Comput Struct Biotechnol J. 2022 Jul 6;20:3567-3580. doi: 10.1016/j.csbj.2022.07.005. eCollection 2022. Comput Struct Biotechnol J. 2022. PMID: 35860410 Free PMC article.
-
Prevalence and architecture of posttranscriptionally impaired synonymous mutations in 8,320 genomes across 22 cancer types.Nucleic Acids Res. 2020 Feb 20;48(3):1192-1205. doi: 10.1093/nar/gkaa019. Nucleic Acids Res. 2020. PMID: 31950163 Free PMC article.
-
SomaMutDB: a database of somatic mutations in normal human tissues.Nucleic Acids Res. 2022 Jan 7;50(D1):D1100-D1108. doi: 10.1093/nar/gkab914. Nucleic Acids Res. 2022. PMID: 34634815 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

