Genomic distribution of a novel Pyrenophora tritici-repentis ToxA insertion element
- PMID: 30379913
- PMCID: PMC6209302
- DOI: 10.1371/journal.pone.0206586
Genomic distribution of a novel Pyrenophora tritici-repentis ToxA insertion element
Abstract
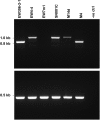
The ToxA effector is a major virulence gene of Pyrenophora tritici-repentis (Ptr), a necrotrophic fungus and the causal agent of tan spot disease of wheat. ToxA and co-located genes are believed to be the result of a recent horizontally transferred highly conserved 14kb region a major pathogenic event for Ptr. Since this event, monitoring isolates for pathogenic changes has become important to help understand the underlying mechanisms in play. Here we examined ToxA in 100 Ptr isolates from Australia, Europe, North and South America and the Middle East, and uncovered in isolates from Denmark, Germany and New Zealand a new variation, a novel 166 bp insertion element (PtrHp1) which can form a perfectly matched 59 bp inverted repeat hairpin structure located downstream of the ToxA coding sequence in the 3' UTR exon. A wider examination revealed PtrHp1 elements to be distributed throughout the genome. Analysis of genomes from Australia and North America had 50-112 perfect copies that often overlap other genes. The hairpin element appears to be unique to Ptr and the lack of ancient origins in other species suggests that PtrHp1 emerged after Ptr speciation. Furthermore, the ToxA UTR insertion site is identical for different isolates, which suggests a single insertion event occurred after the ToxA horizontal transfer. In vitro and in planta-detached leaf assays found that the PtrHp1 element insertion had no effect on ToxA expression. However, variation in the expression of ToxA was detected between the Ptr isolates from different demographic locations, which appears to be unrelated to the presence of the element. We envision that this discovery may contribute towards future understanding of the possible role of hairpin elements in Ptr.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
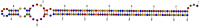


Similar articles
-
Transposon-Mediated Horizontal Transfer of the Host-Specific Virulence Protein ToxA between Three Fungal Wheat Pathogens.mBio. 2019 Sep 10;10(5):e01515-19. doi: 10.1128/mBio.01515-19. mBio. 2019. PMID: 31506307 Free PMC article.
-
The pangenome of the wheat pathogen Pyrenophora tritici-repentis reveals novel transposons associated with necrotrophic effectors ToxA and ToxB.BMC Biol. 2022 Oct 24;20(1):239. doi: 10.1186/s12915-022-01433-w. BMC Biol. 2022. PMID: 36280878 Free PMC article.
-
Homologs of ToxB, a host-selective toxin gene from Pyrenophora tritici-repentis, are present in the genome of sister-species Pyrenophora bromi and other members of the Ascomycota.Fungal Genet Biol. 2008 Mar;45(3):363-77. doi: 10.1016/j.fgb.2007.10.014. Epub 2007 Oct 30. Fungal Genet Biol. 2008. PMID: 18226934
-
Host-selective toxins, Ptr ToxA and Ptr ToxB, as necrotrophic effectors in the Pyrenophora tritici-repentis-wheat interaction.New Phytol. 2010 Sep;187(4):911-9. doi: 10.1111/j.1469-8137.2010.03362.x. Epub 2010 Jul 14. New Phytol. 2010. PMID: 20646221 Review.
-
Genetics of tan spot resistance in wheat.Theor Appl Genet. 2013 Sep;126(9):2197-217. doi: 10.1007/s00122-013-2157-y. Epub 2013 Jul 25. Theor Appl Genet. 2013. PMID: 23884599 Review.
Cited by
-
The first genome assembly of fungal pathogen Pyrenophora tritici-repentis race 1 isolate using Oxford Nanopore MinION sequencing.BMC Res Notes. 2021 Aug 28;14(1):334. doi: 10.1186/s13104-021-05751-0. BMC Res Notes. 2021. PMID: 34454585 Free PMC article.
-
Fungal Effectoromics: A World in Constant Evolution.Int J Mol Sci. 2022 Nov 3;23(21):13433. doi: 10.3390/ijms232113433. Int J Mol Sci. 2022. PMID: 36362218 Free PMC article. Review.
-
Categorization of Orthologous Gene Clusters in 92 Ascomycota Genomes Reveals Functions Important for Phytopathogenicity.J Fungi (Basel). 2021 Apr 27;7(5):337. doi: 10.3390/jof7050337. J Fungi (Basel). 2021. PMID: 33925458 Free PMC article.
-
Research advances in the Pyrenophora teres-barley interaction.Mol Plant Pathol. 2020 Feb;21(2):272-288. doi: 10.1111/mpp.12896. Epub 2019 Dec 13. Mol Plant Pathol. 2020. PMID: 31837102 Free PMC article. Review.
-
Population-level transposable element expression dynamics influence trait evolution in a fungal crop pathogen.mBio. 2024 Mar 13;15(3):e0284023. doi: 10.1128/mbio.02840-23. Epub 2024 Feb 13. mBio. 2024. PMID: 38349152 Free PMC article.
References
-
- Moffat C, Santana MF. Diseases affecting wheat: tan spot In: O R, editor. Integrated disease management of wheat and barley: Burleigh dodds; 2018.
-
- Tan KC, Oliver RP, Solomon PS, Moffat CS. Proteinaceous necrotrophic effectors in fungal virulence. Funct Plant Biol. 2010;37(10):907–12. 10.1071/Fp10067 - DOI
-
- Tomas A, Feng GH, Reeck GR, Bockus WW, Leach JE. Purification of a Cultivar-Specific Toxin from Pyrenophora-Tritici-Repentis, Causal Agent of Tan Spot of Wheat. Mol Plant Microbe In. 1990;3(4):221–4. 10.1094/Mpmi-3-221 - DOI
-
- Tuori RP, Wolpert TJ, Ciuffetti LM. Purification and immunological characterization of toxic components from cultures of Pyrenophora tritici-repentis. Mol Plant Microbe Interact. 1995;8(1):41–8. . - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

