Appropriate Assignment of Fossil Calibration Information Minimizes the Difference between Phylogenetic and Pedigree Mutation Rates in Humans
- PMID: 30360410
- PMCID: PMC6316143
- DOI: 10.3390/life8040049
Appropriate Assignment of Fossil Calibration Information Minimizes the Difference between Phylogenetic and Pedigree Mutation Rates in Humans
Abstract
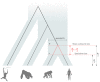
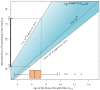
Studies that measured mutation rates in human populations using pedigrees have reported values that differ significantly from rates estimated from the phylogenetic comparison of humans and chimpanzees. Consequently, exchanges between mutation rate values across different timescales lead to conflicting divergence time estimates. It has been argued that this variation of mutation rate estimates across hominoid evolution is in part caused by incorrect assignment of calibration information to the mean coalescent time among loci, instead of the true genetic isolation (speciation) time between humans and chimpanzees. In this study, we investigated the feasibility of estimating the human pedigree mutation rate using phylogenetic data from the genomes of great apes. We found that, when calibration information was correctly assigned to the human⁻chimpanzee speciation time (and not to the coalescent time), estimates of phylogenetic mutation rates were statistically equivalent to the estimates previously reported using studies of human pedigrees. We conclude that, within the range of biologically realistic ancestral generation times, part of the difference between whole-genome phylogenetic and pedigree mutation rates is due to inappropriate assignment of fossil calibration information to the mean coalescent time instead of the speciation time. Although our results focus on the human⁻chimpanzee divergence, our findings are general, and relevant to the inference of the timescale of the tree of life.
Keywords: coalescent; generation time; primate evolution; speciation; species tree.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Estimation of hominoid ancestral population sizes under bayesian coalescent models incorporating mutation rate variation and sequencing errors.Mol Biol Evol. 2008 Sep;25(9):1979-94. doi: 10.1093/molbev/msn148. Epub 2008 Jul 4. Mol Biol Evol. 2008. PMID: 18603620
-
Fossil calibration of molecular divergence infers a moderate mutation rate and recent radiations for pinus.Mol Biol Evol. 2007 Jan;24(1):90-101. doi: 10.1093/molbev/msl131. Epub 2006 Sep 22. Mol Biol Evol. 2007. PMID: 16997907
-
New evidence for the recent divergence of Devil's Hole pupfish and the plausibility of elevated mutation rates in endangered taxa.Mol Ecol. 2018 Feb;27(4):831-838. doi: 10.1111/mec.14404. Epub 2017 Nov 28. Mol Ecol. 2018. PMID: 29148600
-
Catarrhine primate divergence dates estimated from complete mitochondrial genomes: concordance with fossil and nuclear DNA evidence.J Hum Evol. 2005 Mar;48(3):237-57. doi: 10.1016/j.jhevol.2004.11.007. Epub 2005 Jan 20. J Hum Evol. 2005. PMID: 15737392 Review.
-
Time-dependent rates of molecular evolution.Mol Ecol. 2011 Aug;20(15):3087-101. doi: 10.1111/j.1365-294X.2011.05178.x. Epub 2011 Jul 8. Mol Ecol. 2011. PMID: 21740474 Review.
Cited by
-
Data partitioning and correction for ascertainment bias reduce the uncertainty of placental mammal divergence times inferred from the morphological clock.Ecol Evol. 2019 Jan 30;9(4):2255-2262. doi: 10.1002/ece3.4921. eCollection 2019 Feb. Ecol Evol. 2019. PMID: 30847109 Free PMC article.
-
The Estimated Pacemaker for Great Apes Supports the Hominoid Slowdown Hypothesis.Evol Bioinform Online. 2019 Jun 13;15:1176934319855988. doi: 10.1177/1176934319855988. eCollection 2019. Evol Bioinform Online. 2019. PMID: 31223232 Free PMC article.
References
-
- Kimura M. The Neutral Theory of Molecular Evolution. Cambridge University Press; New York, CA, USA: 1983.
-
- Nei M. Mutation-Driven Evolution. Oxford University Press; New York, CA, USA: 2013.
-
- Nei M., Kumar S. Molecular Evolution and Phylogenetics. Oxford University Press; New York, CA, USA: 2000.
Grants and funding
LinkOut - more resources
Full Text Sources

