BCL6 Inhibitor-Mediated Downregulation of Phosphorylated SAMHD1 and T Cell Activation Are Associated with Decreased HIV Infection and Reactivation
- PMID: 30355686
- PMCID: PMC6321929
- DOI: 10.1128/JVI.01073-18
BCL6 Inhibitor-Mediated Downregulation of Phosphorylated SAMHD1 and T Cell Activation Are Associated with Decreased HIV Infection and Reactivation
Abstract
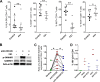
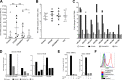
Clearance of HIV-infected germinal center (GC) CD4+ follicular helper T cells (Tfh) after combination antiretroviral therapy (ART) is essential to an HIV cure. Blocking B cell lymphoma 6 (BCL6; the master transcription factor for Tfh cells) represses HIV infection of tonsillar CD4+ Tfh ex vivo, reduces GC formation, and limits immune activation in vivo We assessed the anti-HIV activity of a novel BCL6 inhibitor, FX1, in Tfh/non-Tfh CD4+ T cells and its impact on T cell activation and SAMHD1 phosphorylation (Thr592). FX1 repressed HIV-1 infection of peripheral CD4+ T cells and tonsillar Tfh/non-Tfh CD4+ T cells (P < 0.05) and total elongated and multispliced HIV-1 RNA production during the first round of viral life cycle (P < 0.01). Using purified circulating CD4+ T cells from uninfected donors, we demonstrate that FX1 treatment resulted in downregulation pSAMHD1 expression (P < 0.05) and T cell activation (HLA-DR, CD25, and Ki67; P < 0.01) ex vivo corresponding with inhibition of HIV-1 and HIV-2 replication. Ex vivo HIV-1 reactivation using purified peripheral CD4+ T cells from HIV-infected ART-suppressed donors was also blocked by FX1 treatment (P < 0.01). Our results indicate that BCL6 function contributes to Tfh/non-Tfh CD4+ T cell activation and cellular susceptibility to HIV infection. BCL6 inhibition represents a novel therapeutic strategy to potentiate HIV suppression in Tfh/non-Tfh CD4+ T cells without reactivation of latent virus.IMPORTANCE The expansion and accumulation of HIV-infected BCL6+ Tfh CD4+ T cells are thought to contribute to the persistence of viral reservoirs in infected subjects undergoing ART. Two mechanisms have been raised for the preferential retention of HIV within Tfh CD4+ T cells: (i) antiretroviral drugs have limited tissue distribution, resulting in insufficient tissue concentration and lower efficacy in controlling HIV replication in lymphoid tissues, and (ii) cytotoxic CD8+ T cells within lymphoid tissues express low levels of chemokine receptor (CXCR5), thus limiting their ability to enter the GCs to control/eliminate HIV-infected Tfh cells. Our results indicate that the BCL6 inhibitor FX1 can not only repress HIV infection of tonsillar Tfh ex vivo but also suppress HIV infection and reactivation in primary, non-Tfh CD4+ T cells. Our study provides a rationale for targeting BCL6 protein to extend ART-mediated reduction of persistent HIV and/or support strategies toward HIV remission beyond ART cessation.
Keywords: BCL6; HIV infection; SAMHD1; T cell activation; Tfh.
Copyright © 2019 American Society for Microbiology.
Figures



Similar articles
-
BCL6 BTB-specific inhibitor reversely represses T-cell activation, Tfh cells differentiation, and germinal center reaction in vivo.Eur J Immunol. 2021 Oct;51(10):2441-2451. doi: 10.1002/eji.202049150. Epub 2021 Sep 16. Eur J Immunol. 2021. PMID: 34287839 Free PMC article.
-
BCL6 BTB-specific inhibition via FX1 treatment reduces Tfh cells and reverses lymphoid follicle hyperplasia in Indian rhesus macaque (Macaca mulatta).J Med Primatol. 2020 Feb;49(1):26-33. doi: 10.1111/jmp.12438. Epub 2019 Oct 1. J Med Primatol. 2020. PMID: 31571234 Free PMC article.
-
Small-molecule BCL6 inhibitor protects chronic cardiac transplant rejection and inhibits T follicular helper cell expansion and humoral response.Front Pharmacol. 2023 Mar 17;14:1140703. doi: 10.3389/fphar.2023.1140703. eCollection 2023. Front Pharmacol. 2023. PMID: 37007047 Free PMC article.
-
Divergent Expression of CXCR5 and CCR5 on CD4+ T Cells and the Paradoxical Accumulation of T Follicular Helper Cells during HIV Infection.Front Immunol. 2017 May 12;8:495. doi: 10.3389/fimmu.2017.00495. eCollection 2017. Front Immunol. 2017. PMID: 28553284 Free PMC article. Review.
-
T Cell Subsets in the Germinal Center: Lessons from the Macaque Model.Front Immunol. 2018 Feb 26;9:348. doi: 10.3389/fimmu.2018.00348. eCollection 2018. Front Immunol. 2018. PMID: 29535724 Free PMC article. Review.
Cited by
-
Therapeutic melanoma vaccine with cancer stem cell phenotype represses exhaustion and maintains antigen-specific T cell stemness by up-regulating BCL6.Oncoimmunology. 2020 Jan 11;9(1):1710063. doi: 10.1080/2162402X.2019.1710063. eCollection 2020. Oncoimmunology. 2020. PMID: 32002306 Free PMC article.
-
Hepatitis B and HIV-1 2019 IAS Cure Forum: lessons and benefits from interdisciplinary research.J Virus Erad. 2019 Nov 4;5(4):234-244. doi: 10.1016/S2055-6640(20)30027-3. J Virus Erad. 2019. PMID: 31754448 Free PMC article. No abstract available.
-
Recommendations for measuring HIV reservoir size in cure-directed clinical trials.Nat Med. 2020 Sep;26(9):1339-1350. doi: 10.1038/s41591-020-1022-1. Epub 2020 Sep 7. Nat Med. 2020. PMID: 32895573 Free PMC article. Review.
-
Integrated Assessment of Viral Transcription, Antigen Presentation, and CD8+ T Cell Function Reveals Multiple Limitations of Class I-Selective Histone Deacetylase Inhibitors during HIV-1 Latency Reversal.J Virol. 2020 Apr 16;94(9):e01845-19. doi: 10.1128/JVI.01845-19. Print 2020 Apr 16. J Virol. 2020. PMID: 32051267 Free PMC article.
-
Are HIV-1-Specific Antibody Levels Potentially Useful Laboratory Markers to Estimate HIV Reservoir Size? A Review.Front Immunol. 2021 Nov 11;12:786341. doi: 10.3389/fimmu.2021.786341. eCollection 2021. Front Immunol. 2021. PMID: 34858439 Free PMC article. Review.
References
-
- Ruffin N, Brezar V, Ayinde D, Lefebvre C, Schulze Zur Wiesch J, van Lunzen J, Bockhorn M, Schwartz O, Hocini H, Lelievre JD, Banchereau J, Levy Y, Seddiki N. 2015. Low SAMHD1 expression following T-cell activation and proliferation renders CD4+ T cells susceptible to HIV-1. AIDS 29:519–530. doi:10.1097/QAD.0000000000000594. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

