Peyer's patch myeloid cells infection by Listeria signals through gp38+ stromal cells and locks intestinal villus invasion
- PMID: 30355616
- PMCID: PMC6219733
- DOI: 10.1084/jem.20181210
Peyer's patch myeloid cells infection by Listeria signals through gp38+ stromal cells and locks intestinal villus invasion
Abstract
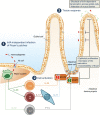
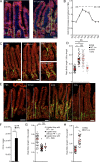
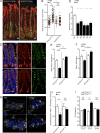
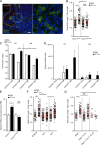
The foodborne pathogen Listeria monocytogenes (Lm) crosses the intestinal villus epithelium via goblet cells (GCs) upon the interaction of Lm surface protein InlA with its receptor E-cadherin. Here, we show that Lm infection accelerates intestinal villus epithelium renewal while decreasing the number of GCs expressing luminally accessible E-cadherin, thereby locking Lm portal of entry. This novel innate immune response to an enteropathogen is triggered by the infection of Peyer's patch CX3CR1+ cells and the ensuing production of IL-23. It requires STAT3 phosphorylation in epithelial cells in response to IL-22 and IL-11 expressed by lamina propria gp38+ stromal cells. Lm-induced IFN-γ signaling and STAT1 phosphorylation in epithelial cells is also critical for Lm-associated intestinal epithelium response. GC depletion also leads to a decrease in colon mucus barrier thickness, thereby increasing host susceptibility to colitis. This study unveils a novel innate immune response to an enteropathogen, which implicates gp38+ stromal cells and locks intestinal villus invasion, but favors colitis.
© 2018 Disson et al.
Figures
Similar articles
-
Functional genomic studies of the intestinal response to a foodborne enteropathogen in a humanized gnotobiotic mouse model.J Biol Chem. 2007 May 18;282(20):15065-72. doi: 10.1074/jbc.M610926200. Epub 2007 Mar 27. J Biol Chem. 2007. PMID: 17389602
-
PI3-kinase activation is critical for host barrier permissiveness to Listeria monocytogenes.J Exp Med. 2015 Feb 9;212(2):165-83. doi: 10.1084/jem.20141406. Epub 2015 Jan 26. J Exp Med. 2015. PMID: 25624443 Free PMC article.
-
A microbiological, histopathological and immunohistological study of the intragastric inoculation of Listeria monocytogenes in mice.J Comp Pathol. 1992 Jul;107(1):1-9. doi: 10.1016/0021-9975(92)90090-h. J Comp Pathol. 1992. PMID: 1430342
-
Entry of microbes into the host: using M cells to break the mucosal barrier.Curr Opin Immunol. 1995 Aug;7(4):474-8. doi: 10.1016/0952-7915(95)80091-3. Curr Opin Immunol. 1995. PMID: 7495510 Review.
-
Intestinal M cells and their role in bacterial infection.Int J Med Microbiol. 2003 Apr;293(1):17-39. doi: 10.1078/1438-4221-00242. Int J Med Microbiol. 2003. PMID: 12755364 Review.
Cited by
-
Innate and Adaptive Immune Responses during Listeria monocytogenes Infection.Microbiol Spectr. 2019 May;7(3):10.1128/microbiolspec.gpp3-0065-2019. doi: 10.1128/microbiolspec.GPP3-0065-2019. Microbiol Spectr. 2019. PMID: 31124430 Free PMC article.
-
Trained ILC3 responses promote intestinal defense.Science. 2022 Feb 25;375(6583):859-863. doi: 10.1126/science.aaz8777. Epub 2022 Feb 24. Science. 2022. PMID: 35201883 Free PMC article.
-
Fibroblastic reticular cells at the nexus of innate and adaptive immune responses.Immunol Rev. 2019 May;289(1):31-41. doi: 10.1111/imr.12748. Immunol Rev. 2019. PMID: 30977192 Free PMC article. Review.
-
Human Listeriosis.Clin Microbiol Rev. 2023 Mar 23;36(1):e0006019. doi: 10.1128/cmr.00060-19. Epub 2022 Dec 8. Clin Microbiol Rev. 2023. PMID: 36475874 Free PMC article. Review.
-
Lymph node stromal cells vary in susceptibility to infection but can support the intracellular growth of Listeria monocytogenes.J Leukoc Biol. 2024 Jun 28;116(1):132-145. doi: 10.1093/jleuko/qiae040. J Leukoc Biol. 2024. PMID: 38416405 Free PMC article.
References
-
- Andersson A., Dai W.J., Di Santo J.P., and Brombacher F.. 1998. Early IFN-gamma production and innate immunity during Listeria monocytogenes infection in the absence of NK cells. J. Immunol. 161:5600–5606. - PubMed
-
- Andoh A., Zhang Z., Inatomi O., Fujino S., Deguchi Y., Araki Y., Tsujikawa T., Kitoh K., Kim-Mitsuyama S., Takayanagi A., et al. . 2005. Interleukin-22, a member of the IL-10 subfamily, induces inflammatory responses in colonic subepithelial myofibroblasts. Gastroenterology. 129:969–984. 10.1053/j.gastro.2005.06.071 - DOI - PubMed
-
- Aparicio-Domingo P., Romera-Hernandez M., Karrich J.J., Cornelissen F., Papazian N., Lindenbergh-Kortleve D.J., Butler J.A., Boon L., Coles M.C., Samsom J.N., and Cupedo T.. 2015. Type 3 innate lymphoid cells maintain intestinal epithelial stem cells after tissue damage. J. Exp. Med. 212:1783–1791. 10.1084/jem.20150318 - DOI - PMC - PubMed
-
- Asker N., Axelsson M.A., Olofsson S.O., and Hansson G.C.. 1998. Dimerization of the human MUC2 mucin in the endoplasmic reticulum is followed by a N-glycosylation-dependent transfer of the mono- and dimers to the Golgi apparatus. J. Biol. Chem. 273:18857–18863. 10.1074/jbc.273.30.18857 - DOI - PubMed
-
- Aychek T., Mildner A., Yona S., Kim K.W., Lampl N., Reich-Zeliger S., Boon L., Yogev N., Waisman A., Cua D.J., and Jung S.. 2015. IL-23-mediated mononuclear phagocyte crosstalk protects mice from Citrobacter rodentium-induced colon immunopathology. Nat. Commun. 6:6525 10.1038/ncomms7525 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

