Long non-coding RNA ChRO1 facilitates ATRX/DAXX-dependent H3.3 deposition for transcription-associated heterochromatin reorganization
- PMID: 30335163
- PMCID: PMC6294499
- DOI: 10.1093/nar/gky923
Long non-coding RNA ChRO1 facilitates ATRX/DAXX-dependent H3.3 deposition for transcription-associated heterochromatin reorganization
Abstract
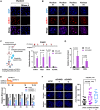
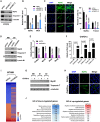
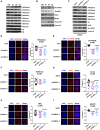
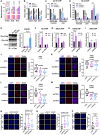
Constitutive heterochromatin undergoes a dynamic clustering and spatial reorganization during myogenic differentiation. However the detailed mechanisms and its role in cell differentiation remain largely elusive. Here, we report the identification of a muscle-specific long non-coding RNA, ChRO1, involved in constitutive heterochromatin reorganization. ChRO1 is induced during terminal differentiation of myoblasts, and is specifically localized to the chromocenters in myotubes. ChRO1 is required for efficient cell differentiation, with global impacts on gene expression. It influences DNA methylation and chromatin compaction at peri/centromeric regions. Inhibition of ChRO1 leads to defects in the spatial fusion of chromocenters, and mislocalization of H4K20 trimethylation, Suv420H2, HP1, MeCP2 and cohesin. In particular, ChRO1 specifically associates with ATRX/DAXX/H3.3 complex at chromocenters to promote H3.3 incorporation and transcriptional induction of satellite repeats, which is essential for chromocenter clustering. Thus, our results unveil a mechanism involving a lncRNA that plays a role in large-scale heterochromatin reorganization and cell differentiation.
Figures

Similar articles
-
PML protein organizes heterochromatin domains where it regulates histone H3.3 deposition by ATRX/DAXX.Genome Res. 2017 Jun;27(6):913-921. doi: 10.1101/gr.215830.116. Epub 2017 Mar 24. Genome Res. 2017. PMID: 28341773 Free PMC article.
-
ATRX Contributes to MeCP2-Mediated Pericentric Heterochromatin Organization during Neural Differentiation.Int J Mol Sci. 2019 Oct 29;20(21):5371. doi: 10.3390/ijms20215371. Int J Mol Sci. 2019. PMID: 31671722 Free PMC article.
-
Methyl CpG-binding proteins induce large-scale chromatin reorganization during terminal differentiation.J Cell Biol. 2005 Jun 6;169(5):733-43. doi: 10.1083/jcb.200502062. J Cell Biol. 2005. PMID: 15939760 Free PMC article.
-
New players in heterochromatin silencing: histone variant H3.3 and the ATRX/DAXX chaperone.Nucleic Acids Res. 2016 Feb 29;44(4):1496-501. doi: 10.1093/nar/gkw012. Epub 2016 Jan 14. Nucleic Acids Res. 2016. PMID: 26773061 Free PMC article. Review.
-
Maintaining memory of silencing at imprinted differentially methylated regions.Cell Mol Life Sci. 2016 May;73(9):1871-9. doi: 10.1007/s00018-016-2157-6. Epub 2016 Feb 16. Cell Mol Life Sci. 2016. PMID: 26883803 Free PMC article. Review.
Cited by
-
Neurodevelopmental Disorders Caused by Defective Chromatin Remodeling: Phenotypic Complexity Is Highlighted by a Review of ATRX Function.Front Genet. 2020 Aug 11;11:885. doi: 10.3389/fgene.2020.00885. eCollection 2020. Front Genet. 2020. PMID: 32849845 Free PMC article. Review.
-
Long Non-coding RNAs (lncRNAs), A New Target in Stroke.Cell Mol Neurobiol. 2022 Apr;42(3):501-519. doi: 10.1007/s10571-020-00954-8. Epub 2020 Aug 31. Cell Mol Neurobiol. 2022. PMID: 32865676 Review.
-
Single-genome analysis reveals a heterogeneous association of the herpes simplex virus genome with H3K27me2 and the reader PHF20L1 following infection of human fibroblasts.mBio. 2024 Apr 10;15(4):e0327823. doi: 10.1128/mbio.03278-23. Epub 2024 Feb 27. mBio. 2024. PMID: 38411116 Free PMC article.
-
DAXX adds a de novo H3.3K9me3 deposition pathway to the histone chaperone network.Mol Cell. 2023 Apr 6;83(7):1075-1092.e9. doi: 10.1016/j.molcel.2023.02.009. Epub 2023 Mar 2. Mol Cell. 2023. PMID: 36868228 Free PMC article.
-
Epigenetic Factors That Control Pericentric Heterochromatin Organization in Mammals.Genes (Basel). 2020 May 28;11(6):595. doi: 10.3390/genes11060595. Genes (Basel). 2020. PMID: 32481609 Free PMC article. Review.
References
-
- Solovei I., Wang A.S., Thanisch K., Schmidt C.S., Krebs S., Zwerger M., Cohen T.V., Devys D., Foisner R., Peichl L. et al. . LBR and lamin A/C sequentially tether peripheral heterochromatin and inversely regulate differentiation. Cell. 2013; 152:584–598. - PubMed
-
- Padeken J., Mendiburo M.J., Chlamydas S., Schwarz H.J., Kremmer E., Heun P.. The nucleoplasmin homolog NLP mediates centromere clustering and anchoring to the nucleolus. Mol. Cell. 2013; 50:236–249. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

