The Emerging Role of DNA Damage in the Pathogenesis of the C9orf72 Repeat Expansion in Amyotrophic Lateral Sclerosis
- PMID: 30322030
- PMCID: PMC6213462
- DOI: 10.3390/ijms19103137
The Emerging Role of DNA Damage in the Pathogenesis of the C9orf72 Repeat Expansion in Amyotrophic Lateral Sclerosis
Abstract
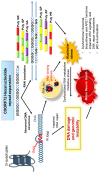
Amyotrophic lateral sclerosis (ALS) is a fatal, rapidly progressing neurodegenerative disease affecting motor neurons, and frontotemporal dementia (FTD) is a behavioural disorder resulting in early-onset dementia. Hexanucleotide (G4C2) repeat expansions in the gene encoding chromosome 9 open reading frame 72 (C9orf72) are the major cause of familial forms of both ALS (~40%) and FTD (~20%) worldwide. The C9orf72 repeat expansion is known to form abnormal nuclei acid structures, such as hairpins, G-quadruplexes, and R-loops, which are increasingly associated with human diseases involving microsatellite repeats. These configurations form during normal cellular processes, but if they persist they also damage DNA, and hence are a serious threat to genome integrity. It is unclear how the repeat expansion in C9orf72 causes ALS, but recent evidence implicates DNA damage in neurodegeneration. This may arise from abnormal nucleic acid structures, the greatly expanded C9orf72 RNA, or by repeat-associated non-ATG (RAN) translation, which generates toxic dipeptide repeat proteins. In this review, we detail recent advances implicating DNA damage in C9orf72-ALS. Furthermore, we also discuss increasing evidence that targeting these aberrant C9orf72 confirmations may have therapeutic value for ALS, thus revealing new avenues for drug discovery for this disorder.
Keywords: ALS; C9orf72; R loops, nucleolar stress; motor neuron disease; neurodegeneration.
Conflict of interest statement
The authors have no conflict of interest to declare.
Figures

Similar articles
-
The DNA damage response (DDR) is induced by the C9orf72 repeat expansion in amyotrophic lateral sclerosis.Hum Mol Genet. 2017 Aug 1;26(15):2882-2896. doi: 10.1093/hmg/ddx170. Hum Mol Genet. 2017. PMID: 28481984
-
Molecular Mechanisms of Neurodegeneration Related to C9orf72 Hexanucleotide Repeat Expansion.Behav Neurol. 2019 Jan 15;2019:2909168. doi: 10.1155/2019/2909168. eCollection 2019. Behav Neurol. 2019. PMID: 30774737 Free PMC article. Review.
-
C9orf72 ALS-FTD: recent evidence for dysregulation of the autophagy-lysosome pathway at multiple levels.Autophagy. 2021 Nov;17(11):3306-3322. doi: 10.1080/15548627.2021.1872189. Epub 2021 Feb 26. Autophagy. 2021. PMID: 33632058 Free PMC article. Review.
-
All in the Family: Repeats and ALS/FTD.Trends Neurosci. 2018 May;41(5):247-250. doi: 10.1016/j.tins.2018.03.010. Trends Neurosci. 2018. PMID: 29703376 Free PMC article. Review.
-
Insights into the pathogenic mechanisms of Chromosome 9 open reading frame 72 (C9orf72) repeat expansions.J Neurochem. 2016 Aug;138 Suppl 1:145-62. doi: 10.1111/jnc.13623. Epub 2016 Jun 15. J Neurochem. 2016. PMID: 27016280 Review.
Cited by
-
Chronological and Biological Aging in Amyotrophic Lateral Sclerosis and the Potential of Senolytic Therapies.Cells. 2024 May 28;13(11):928. doi: 10.3390/cells13110928. Cells. 2024. PMID: 38891059 Free PMC article. Review.
-
Neuronal STING activation in amyotrophic lateral sclerosis and frontotemporal dementia.Acta Neuropathol. 2024 Mar 13;147(1):56. doi: 10.1007/s00401-024-02688-z. Acta Neuropathol. 2024. PMID: 38478117 Free PMC article.
-
Emerging Evidence Highlighting the Importance of Redox Dysregulation in the Pathogenesis of Amyotrophic Lateral Sclerosis (ALS).Front Cell Neurosci. 2021 Feb 18;14:581950. doi: 10.3389/fncel.2020.581950. eCollection 2020. Front Cell Neurosci. 2021. PMID: 33679322 Free PMC article.
-
Senataxin deficiency disrupts proteostasis through nucleolar ncRNA-driven protein aggregation.J Cell Biol. 2024 Jul 1;223(7):e202309036. doi: 10.1083/jcb.202309036. Epub 2024 May 8. J Cell Biol. 2024. PMID: 38717338 Free PMC article.
-
Motor Neuron Susceptibility in ALS/FTD.Front Neurosci. 2019 Jun 27;13:532. doi: 10.3389/fnins.2019.00532. eCollection 2019. Front Neurosci. 2019. PMID: 31316328 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

