ARSENATE INDUCED CHLOROSIS 1/ TRANSLOCON AT THE OUTER ENVOLOPE MEMBRANE OF CHLOROPLASTS 132 Protects Chloroplasts from Arsenic Toxicity
- PMID: 30309965
- PMCID: PMC6288752
- DOI: 10.1104/pp.18.01042
ARSENATE INDUCED CHLOROSIS 1/ TRANSLOCON AT THE OUTER ENVOLOPE MEMBRANE OF CHLOROPLASTS 132 Protects Chloroplasts from Arsenic Toxicity
Abstract
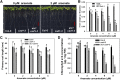
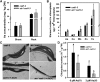
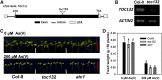
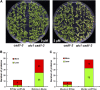
Arsenic (As) is highly toxic to plants and detoxified primarily through complexation with phytochelatins (PCs) and other thiol compounds. To understand the mechanisms of As toxicity and detoxification beyond PCs, we isolated an arsenate-sensitive mutant of Arabidopsis (Arabidopsis thaliana), arsenate induced chlorosis1 (aic1), in the background of the PC synthase-defective mutant cadmium-sensitive1-3 (cad1-3). Under arsenate stress, aic1 cad1-3 showed larger decreases in chlorophyll content and the number and size of chloroplasts than cad1-3 and a severely distorted chloroplast structure. The aic1 single mutant also was more sensitive to arsenate than the wild type (Columbia-0). As concentrations in the roots, shoots, and chloroplasts were similar between aic1 cad1-3 and cad1-3 Using genome resequencing and complementation, TRANSLOCON AT THE OUTER ENVOLOPE MEMBRANE OF CHLOROPLAST132 (TOC132) was identified as the mutant gene, which encodes a translocon protein involved in the import of preproteins from the cytoplasm into the chloroplasts. Proteomic analysis showed that the proteome of aic1 cad1-3 chloroplasts was more affected by arsenate stress than that of cad1-3 A number of proteins related to chloroplast ribosomes, photosynthesis, compound synthesis, and thioredoxin systems were less abundant in aic1 cad1-3 than in cad1-3 under arsenate stress. Our results indicate that chloroplasts are a sensitive target of As toxicity and that AIC1/Toc132 plays an important role in protecting chloroplasts from As toxicity.
© 2018 American Society of Plant Biologists. All rights reserved.
Figures


Similar articles
-
Protein phosphatase 2A alleviates cadmium toxicity by modulating ethylene production in Arabidopsis thaliana.Plant Cell Environ. 2020 Apr;43(4):1008-1022. doi: 10.1111/pce.13716. Epub 2020 Jan 28. Plant Cell Environ. 2020. PMID: 31916592
-
Long-distance root-to-shoot transport of phytochelatins and cadmium in Arabidopsis.Proc Natl Acad Sci U S A. 2003 Aug 19;100(17):10118-23. doi: 10.1073/pnas.1734072100. Epub 2003 Aug 8. Proc Natl Acad Sci U S A. 2003. PMID: 12909714 Free PMC article.
-
Mutants impaired in vacuolar metal mobilization identify chloroplasts as a target for cadmium hypersensitivity in Arabidopsis thaliana.Plant Cell Environ. 2013 Apr;36(4):804-17. doi: 10.1111/pce.12016. Epub 2012 Oct 24. Plant Cell Environ. 2013. PMID: 22998565
-
Identification of C-terminal Regions in Arabidopsis thaliana Phytochelatin Synthase 1 Specifically Involved in Activation by Arsenite.Plant Cell Physiol. 2018 Mar 1;59(3):500-509. doi: 10.1093/pcp/pcx204. Plant Cell Physiol. 2018. PMID: 29281059
-
Expression of Arabidopsis phytochelatin synthase 2 is too low to complement an AtPCS1-defective Cad1-3 mutant.Mol Cells. 2005 Feb 28;19(1):81-7. Mol Cells. 2005. PMID: 15750344
Cited by
-
Histological and proteome analyses of Microbacterium foliorum-mediated decrease in arsenic toxicity in Melastoma malabathricum.3 Biotech. 2021 Jul;11(7):336. doi: 10.1007/s13205-021-02864-y. Epub 2021 Jun 16. 3 Biotech. 2021. PMID: 34221807 Free PMC article.
-
Season-dependent physiological behavior of Miscanthus x giganteus growing on heavy-metal contaminated areas in relation to soil properties.Heliyon. 2024 Feb 10;10(4):e25943. doi: 10.1016/j.heliyon.2024.e25943. eCollection 2024 Feb 29. Heliyon. 2024. PMID: 38384526 Free PMC article.
References
-
- Abe A, Kosugi S, Yoshida K, Natsume S, Takagi H, Kanzaki H, Matsumura H, Yoshida K, Mitsuoka C, Tamiru M, et al. (2012) Genome sequencing reveals agronomically important loci in rice using MutMap. Nat Biotechnol 30: 174–178 - PubMed
-
- Baer CD, Edwards JO, Rieger PH (1981) Kinetics of the hydrolysis of arsenate(V) triesters. Inorg Chem 20: 905–907
-
- Bauer J, Chen K, Hiltbunner A, Wehrli E, Eugster M, Schnell D, Kessler F (2000) The major protein import receptor of plastids is essential for chloroplast biogenesis. Nature 403: 203–207 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

