Using Scipion for stream image processing at Cryo-EM facilities
- PMID: 30296492
- PMCID: PMC6303188
- DOI: 10.1016/j.jsb.2018.10.001
Using Scipion for stream image processing at Cryo-EM facilities
Abstract
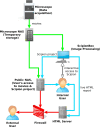
Three dimensional electron microscopy is becoming a very data-intensive field in which vast amounts of experimental images are acquired at high speed. To manage such large-scale projects, we had previously developed a modular workflow system called Scipion (de la Rosa-Trevín et al., 2016). We present here a major extension of Scipion that allows processing of EM images while the data is being acquired. This approach helps to detect problems at early stages, saves computing time and provides users with a detailed evaluation of the data quality before the acquisition is finished. At present, Scipion has been deployed and is in production mode in seven Cryo-EM facilities throughout the world.
Keywords: Electron microscopy; High throughput; Image processing; Live processing; Scipion; Streaming.
Copyright © 2018 Elsevier Inc. All rights reserved.
Figures

Similar articles
-
Using RELION software within the Scipion framework.Acta Crystallogr D Struct Biol. 2021 Apr 1;77(Pt 4):403-410. doi: 10.1107/S2059798321001856. Epub 2021 Mar 30. Acta Crystallogr D Struct Biol. 2021. PMID: 33825701 Free PMC article.
-
A Robust Single-Particle Cryo-Electron Microscopy (cryo-EM) Processing Workflow with cryoSPARC, RELION, and Scipion.J Vis Exp. 2022 Jan 31;(179). doi: 10.3791/63387. J Vis Exp. 2022. PMID: 35104261
-
ScipionTomo: Towards cryo-electron tomography software integration, reproducibility, and validation.J Struct Biol. 2022 Sep;214(3):107872. doi: 10.1016/j.jsb.2022.107872. Epub 2022 Jun 2. J Struct Biol. 2022. PMID: 35660516 Free PMC article.
-
Processing of Structurally Heterogeneous Cryo-EM Data in RELION.Methods Enzymol. 2016;579:125-57. doi: 10.1016/bs.mie.2016.04.012. Epub 2016 May 31. Methods Enzymol. 2016. PMID: 27572726 Review.
-
Combining high throughput and high quality for cryo-electron microscopy data collection.Acta Crystallogr D Struct Biol. 2020 Aug 1;76(Pt 8):724-728. doi: 10.1107/S2059798320008347. Epub 2020 Jul 27. Acta Crystallogr D Struct Biol. 2020. PMID: 32744254 Free PMC article. Review.
Cited by
-
Scipion-ED: a graphical user interface for batch processing and analysis of 3D ED/MicroED data.J Appl Crystallogr. 2022 Apr 22;55(Pt 3):638-646. doi: 10.1107/S1600576722002758. eCollection 2022 Jun 1. J Appl Crystallogr. 2022. PMID: 35719296 Free PMC article.
-
High-resolution structures of malaria parasite actomyosin and actin filaments.PLoS Pathog. 2022 Apr 4;18(4):e1010408. doi: 10.1371/journal.ppat.1010408. eCollection 2022 Apr. PLoS Pathog. 2022. PMID: 35377914 Free PMC article.
-
SIMPLE 3.0. Stream single-particle cryo-EM analysis in real time.J Struct Biol X. 2020 Nov 7;4:100040. doi: 10.1016/j.yjsbx.2020.100040. eCollection 2020. J Struct Biol X. 2020. PMID: 33294840 Free PMC article.
-
CryoEM structure of Drosophila flight muscle thick filaments at 7 Å resolution.Life Sci Alliance. 2020 Jul 27;3(8):e202000823. doi: 10.26508/lsa.202000823. Print 2020 Aug. Life Sci Alliance. 2020. PMID: 32718994 Free PMC article.
-
New tools for automated cryo-EM single-particle analysis in RELION-4.0.Biochem J. 2021 Dec 22;478(24):4169-4185. doi: 10.1042/BCJ20210708. Biochem J. 2021. PMID: 34783343 Free PMC article.
References
-
- Abrishami V., Vargas J., Li X., Cheng Y., Marabini R., Sorzano C.O., Carazo J.M. Alignment of direct detection device micrographs using a robust Optical Flow approach. J. Struct. Biol. 2015;189(3):163–176. - PubMed
-
- Biyani N., Righetto R.D., McLeod R., Caujolle-Bert D., Castano-Diez D., Goldie K.N., Stahlberg H. Focus: the interface between data collection and data processing in cryo-EM. J. Struct. Biol. 2017;198(2):124–133. - PubMed
-
- Clare D.K., Siebert C.A., Hecksel C., Hagen C., Mordhorst V., Grange M., Ashton A.W., Walsh M.A., Grunewald K., Saibil H.R., Stuart D.I., Zhang P. Electron Bio-Imaging Centre (eBIC): the UK national research facility for biological electron microscopy. Acta Crystallogr D Struct. Biol. 2017;73(Pt 6):488–495. - PMC - PubMed
-
- de la Rosa-Trevín J., Quintana A., del Cano L., Zaldívar A., Foche I., Gutiérrez J., Gómez-Blanco J., Burguet-Castell J., Cuenca-Alba J., Abrishami V., Vargas J., Otón J., Sharov G., Vilas J., Navas J., Conesa P., Kazemi M., Marabini R., Sorzano C., Carazo J. Scipion: a software framework toward integration, reproducibility and validation in 3d electron microscopy. J. Struct. Biol. 2016;195(1):93–99. - PubMed
-
- Delageniere S., Brenchereau P., Launer L., Ashton A.W., Leal R., Veyrier S., Gabadinho J., Gordon E.J., Jones S.D., Levik K.E., McSweeney S.M., Monaco S., Nanao M., Spruce D., Svensson O., Walsh M.A., Leonard G.A. ISPyB: an information management system for synchrotron macromolecular crystallography. Bioinformatics. 2011;27(22):3186–3192. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

