Genomic Copy-Number Loss Is Rescued by Self-Limiting Production of DNA Circles
- PMID: 30293780
- PMCID: PMC6214758
- DOI: 10.1016/j.molcel.2018.08.036
Genomic Copy-Number Loss Is Rescued by Self-Limiting Production of DNA Circles
Abstract
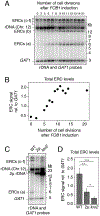
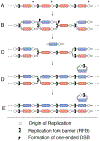
Copy-number changes generate phenotypic variability in health and disease. Whether organisms protect against copy-number changes is largely unknown. Here, we show that Saccharomyces cerevisiae monitors the copy number of its ribosomal DNA (rDNA) and rapidly responds to copy-number loss with the clonal amplification of extrachromosomal rDNA circles (ERCs) from chromosomal repeats. ERC formation is replicative, separable from repeat loss, and reaches a dynamic steady state that responds to the addition of exogenous rDNA copies. ERC levels are also modulated by RNAPI activity and diet, suggesting that rDNA copy number is calibrated against the cellular demand for rRNA. Last, we show that ERCs reinsert into the genome in a dosage-dependent manner, indicating that they provide a reservoir for ultimately increasing rDNA array length. Our results reveal a DNA-based mechanism for rapidly restoring copy number in response to catastrophic gene loss that shares fundamental features with unscheduled copy-number amplifications in cancer cells.
Keywords: Fob1; Hmo1; copy-number variations; eccDNA; genome instability; rDNA; rRNA genes.
Copyright © 2018 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests:
The authors declare no competing interests.
Figures



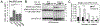

Similar articles
-
DNA replication stress restricts ribosomal DNA copy number.PLoS Genet. 2017 Sep 15;13(9):e1007006. doi: 10.1371/journal.pgen.1007006. eCollection 2017 Sep. PLoS Genet. 2017. PMID: 28915237 Free PMC article.
-
The effect of replication initiation on gene amplification in the rDNA and its relationship to aging.Mol Cell. 2009 Sep 11;35(5):683-93. doi: 10.1016/j.molcel.2009.07.012. Mol Cell. 2009. PMID: 19748361
-
A new method for determining ribosomal DNA copy number shows differences between Saccharomyces cerevisiae populations.Genomics. 2022 Jul;114(4):110430. doi: 10.1016/j.ygeno.2022.110430. Epub 2022 Jul 10. Genomics. 2022. PMID: 35830947
-
How do cells count multi-copy genes?: "Musical Chair" model for preserving the number of rDNA copies.Curr Genet. 2019 Aug;65(4):883-885. doi: 10.1007/s00294-019-00956-0. Epub 2019 Mar 23. Curr Genet. 2019. PMID: 30904990 Review.
-
Regulatory processes that maintain or alter ribosomal DNA stability during the repair of programmed DNA double-strand breaks.Genes Genet Syst. 2023 Sep 30;98(3):103-119. doi: 10.1266/ggs.22-00046. Epub 2022 Aug 4. Genes Genet Syst. 2023. PMID: 35922917 Review.
Cited by
-
Genomic Instability and Epigenetic Changes during Aging.Int J Mol Sci. 2023 Sep 19;24(18):14279. doi: 10.3390/ijms241814279. Int J Mol Sci. 2023. PMID: 37762580 Free PMC article. Review.
-
Glioblastoma: two immune subtypes under the surface of the cold tumor.Aging (Albany NY). 2022 May 23;14(10):4357-4375. doi: 10.18632/aging.204067. Epub 2022 May 23. Aging (Albany NY). 2022. PMID: 35609054 Free PMC article.
-
Small extrachromosomal circular DNA (eccDNA): major functions in evolution and cancer.Mol Cancer. 2021 Sep 3;20(1):113. doi: 10.1186/s12943-021-01413-8. Mol Cancer. 2021. PMID: 34479546 Free PMC article. Review.
-
The long and short of rDNA and yeast replicative aging.Proc Natl Acad Sci U S A. 2022 Jun 7;119(23):e2205124119. doi: 10.1073/pnas.2205124119. Epub 2022 Jun 3. Proc Natl Acad Sci U S A. 2022. PMID: 35658078 Free PMC article. No abstract available.
-
Extrachromosomal circular DNA: biogenesis, structure, functions and diseases.Signal Transduct Target Ther. 2022 Oct 2;7(1):342. doi: 10.1038/s41392-022-01176-8. Signal Transduct Target Ther. 2022. PMID: 36184613 Free PMC article. Review.
References
-
- Albert B, Colleran C, Léger-Silvestre I, Berger AB, Dez C, Normand C, Perez-Fernandez J, McStay B, Gadal O (2013). Structure-function analysis of Hmo1 unveils an ancestral organization of HMG-Box factors involved in ribosomal DNA transcription from yeast to human. Nucleic Acids Res. 41, 10135–10149. doi:10.1093/nar/gkt770 - DOI - PMC - PubMed
-
- Blake D, Luke B, Kanellis P, Jorgensen P, Goh T, Penfold S, Breitkreutz BJ, Durocher D, Peter M, Tyers M (2006). The F-box protein Dia2 overcomes replication impedance to promote genome stability in Saccharomyces cerevisiae. Genetics 174, 1709–1727. doi:10.1534/genetics.106.057836 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

