RAB30 regulates PI4KB (phosphatidylinositol 4-kinase beta)-dependent autophagy against group A Streptococcus
- PMID: 30290718
- PMCID: PMC6351121
- DOI: 10.1080/15548627.2018.1532260
RAB30 regulates PI4KB (phosphatidylinositol 4-kinase beta)-dependent autophagy against group A Streptococcus
Abstract
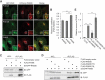
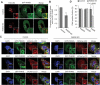
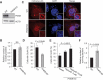
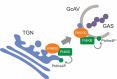
Macroautophagy/autophagy plays an important role in the immune response to invasion by intracellular pathogens such as group A Streptococcus (GAS; Streptococcus pyogenes). We previously identified RAB30, a Golgi-resident GTPase, as a novel anti-bacterial autophagic regulator in the formation of GAS-containing autophagosome-like vacuoles (GcAVs); however, the precise mechanism underlying this process remains elusive. Here, we elucidate a novel property of RAB30: the ability to recruit PI4KB (phosphatidylinositol 4-kinase beta) to the Golgi apparatus and GcAVs. We found that trans-Golgi network (TGN) vesicles were incorporated into GcAVs via RAB30 to promote GcAV formation. Moreover, depletion of phosphatidylinositol-4-phosphate (PtdIns4P), a phosphatidylinositol enriched in the TGN, by wortmannin and phenylarsine oxide, followed by subsequent repletion with exogenous PtdIns4P revealed that PtdIns4P is crucial for GcAV formation. Furthermore, we identify an interaction between RAB30 and PI4KB, in which the knockdown of RAB30 decreased the localization of PI4KB to the TGN and GcAVs. Finally, PI4KB knockout suppressed autophagy by inhibiting GcAV formation, resulting in the increased survival of GAS. Our results demonstrate a novel autophagosomal formation mechanism involving coordinative functions of RAB30 and PI4KB distinct from those utilized in canonical autophagy. Abbreviations: GAS: group A Streptococcus; GcAVs: GAS-containing autophagosome-like vacuoles; PI4KB: phosphatidylinositol 4-kinase beta; PtdIns: phosphatidylinositol; PtdIns3P: phosphatidylinositol-3-phosphate; PtdIns4P: phosphatidylinositol-4-phosphate; PtdIns5P: phosphatidylinositol-5-phosphate; SLO: streptolysin O; TGN: trans-Golgi network; TGOLN2: trans-golgi network protein 2; PH: plekstrin homology; OSBP: oxysterol binding protein.
Keywords: GcAV; PI4KB; group A; trans-Golgi network; xenophagy.
Figures
Similar articles
-
Golgi-Resident GTPase Rab30 Promotes the Biogenesis of Pathogen-Containing Autophagosomes.PLoS One. 2016 Jan 15;11(1):e0147061. doi: 10.1371/journal.pone.0147061. eCollection 2016. PLoS One. 2016. PMID: 26771875 Free PMC article.
-
The STX6-VTI1B-VAMP3 complex facilitates xenophagy by regulating the fusion between recycling endosomes and autophagosomes.Autophagy. 2017 Jan 2;13(1):57-69. doi: 10.1080/15548627.2016.1241924. Epub 2016 Oct 28. Autophagy. 2017. PMID: 27791468 Free PMC article.
-
Group A Streptococcus modulates RAB1- and PIK3C3 complex-dependent autophagy.Autophagy. 2020 Feb;16(2):334-346. doi: 10.1080/15548627.2019.1628539. Epub 2019 Jun 14. Autophagy. 2020. PMID: 31177902 Free PMC article.
-
[Selective autophagy mechanism against Group A Streptococcus infection].Nihon Saikingaku Zasshi. 2018;73(3):193-199. doi: 10.3412/jsb.73.193. Nihon Saikingaku Zasshi. 2018. PMID: 30158393 Review. Japanese.
-
Coordination of Golgi functions by phosphatidylinositol 4-kinases.Trends Cell Biol. 2011 Feb;21(2):113-21. doi: 10.1016/j.tcb.2010.10.002. Epub 2010 Nov 4. Trends Cell Biol. 2011. PMID: 21282087 Free PMC article. Review.
Cited by
-
Golgi-associated Rab GTPases implicated in autophagy.Cell Biosci. 2021 Feb 8;11(1):35. doi: 10.1186/s13578-021-00543-2. Cell Biosci. 2021. PMID: 33557950 Free PMC article. Review.
-
Rab41-mediated ESCRT machinery repairs membrane rupture by a bacterial toxin in xenophagy.Nat Commun. 2023 Oct 6;14(1):6230. doi: 10.1038/s41467-023-42039-2. Nat Commun. 2023. PMID: 37802980 Free PMC article.
-
Proteomic Ligandability Maps of Spirocycle Acrylamide Stereoprobes Identify Covalent ERCC3 Degraders.J Am Chem Soc. 2024 Apr 17;146(15):10393-10406. doi: 10.1021/jacs.3c13448. Epub 2024 Apr 3. J Am Chem Soc. 2024. PMID: 38569115
-
The function of apolipoproteins L (APOLs): relevance for kidney disease, neurotransmission disorders, cancer and viral infection.FEBS J. 2021 Jan;288(2):360-381. doi: 10.1111/febs.15444. Epub 2020 Jun 25. FEBS J. 2021. PMID: 32530132 Free PMC article. Review.
-
Intracellular nanovesicles mediate α5β1 integrin trafficking during cell migration.J Cell Biol. 2021 Oct 4;220(10):e202009028. doi: 10.1083/jcb.202009028. Epub 2021 Jul 21. J Cell Biol. 2021. PMID: 34287617 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
