Rice stripe virus-derived siRNAs play different regulatory roles in rice and in the insect vector Laodelphax striatellus
- PMID: 30286719
- PMCID: PMC6172784
- DOI: 10.1186/s12870-018-1438-7
Rice stripe virus-derived siRNAs play different regulatory roles in rice and in the insect vector Laodelphax striatellus
Abstract
Background: Most plant viruses depend on vector insects for transmission. Upon viral infection, virus-derived small interfering RNAs (vsiRNAs) can target both viral and host transcripts. Rice stripe virus (RSV) is a persistent-propagative virus transmitted by the small brown planthopper (Laodelphax striatellus, Fallen) and can cause a severe disease on rice.
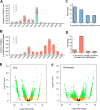
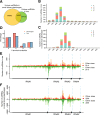
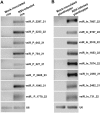
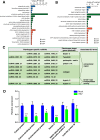
Results: To investigate how vsiRNAs regulate gene expressions in the host plant and the insect vector, we analyzed the expression profiles of small RNAs (sRNAs) and mRNAs in RSV-infected rice and RSV-infected planthopper. We obtained 88,247 vsiRNAs in rice that were predominantly derived from the terminal regions of the RSV RNA segments, and 351,655 vsiRNAs in planthopper that displayed relatively even distributions on RSV RNA segments. 38,112 and 80,698 unique vsiRNAs were found only in rice and planthopper, respectively, while 14,006 unique vsiRNAs were found in both of them. Compared to mock-inoculated rice, 273 genes were significantly down-regulated genes (DRGs) in RSV-infected rice, among which 192 (70.3%) were potential targets of vsiRNAs based on sequence complementarity. Gene ontology (GO) analysis revealed that these 192 DRGs were enriched in genes involved in kinase activity, carbohydrate binding and protein binding. Similarly, 265 DRGs were identified in RSV-infected planthoppers, among which 126 (47.5%) were potential targets of vsiRNAs. These planthopper target genes were enriched in genes that are involved in structural constituent of cuticle, serine-type endopeptidase activity, and oxidoreductase activity.
Conclusions: Taken together, our results reveal that infection by the same virus can generate distinct vsiRNAs in different hosts to potentially regulate different biological processes, thus reflecting distinct virus-host interactions.
Keywords: Rice; Rice stripe virus; Small RNA sequencing; Small brown planthopper; Transcriptome; Virus-derived small interfering RNAs.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures


Similar articles
-
Alternative Splicing Landscape of Small Brown Planthopper and Different Response of JNK2 Isoforms to Rice Stripe Virus Infection.J Virol. 2022 Jan 26;96(2):e0171521. doi: 10.1128/JVI.01715-21. Epub 2021 Nov 10. J Virol. 2022. PMID: 34757837 Free PMC article.
-
Population diversity of rice stripe virus-derived siRNAs in three different hosts and RNAi-based antiviral immunity in Laodelphgax striatellus.PLoS One. 2012;7(9):e46238. doi: 10.1371/journal.pone.0046238. Epub 2012 Sep 28. PLoS One. 2012. PMID: 23029445 Free PMC article.
-
Comparative Transcriptome Analysis of Chemoreception Organs of Laodelphax striatellus in Response to Rice Stripe Virus Infection.Int J Mol Sci. 2021 Sep 24;22(19):10299. doi: 10.3390/ijms221910299. Int J Mol Sci. 2021. PMID: 34638638 Free PMC article.
-
The small brown planthopper (Laodelphax striatellus) as a vector of the rice stripe virus.Arch Insect Biochem Physiol. 2023 Feb;112(2):e21992. doi: 10.1002/arch.21992. Epub 2022 Dec 27. Arch Insect Biochem Physiol. 2023. PMID: 36575628 Review.
-
Recent progress on gene silencing/suppression by virus-derived small interfering RNAs in rice viruses especially Rice grassy stunt virus.Microb Pathog. 2018 Dec;125:210-218. doi: 10.1016/j.micpath.2018.09.021. Epub 2018 Sep 19. Microb Pathog. 2018. PMID: 30243549 Review.
Cited by
-
Interaction between endogenous microRNAs and virus-derived small RNAs controls viral replication in insect vectors.PLoS Pathog. 2022 Jul 7;18(7):e1010709. doi: 10.1371/journal.ppat.1010709. eCollection 2022 Jul. PLoS Pathog. 2022. PMID: 35797383 Free PMC article.
-
Suppression of Rice Stripe Virus Replication in Laodelphax striatellus Using Vector Insect-Derived Double-Stranded RNAs.Plant Pathol J. 2020 Jun 1;36(3):280-288. doi: 10.5423/PPJ.OA.03.2020.0052. Plant Pathol J. 2020. PMID: 32547343 Free PMC article.
-
Alternative Splicing Landscape of Small Brown Planthopper and Different Response of JNK2 Isoforms to Rice Stripe Virus Infection.J Virol. 2022 Jan 26;96(2):e0171521. doi: 10.1128/JVI.01715-21. Epub 2021 Nov 10. J Virol. 2022. PMID: 34757837 Free PMC article.
-
Virus and Viroid-Derived Small RNAs as Modulators of Host Gene Expression: Molecular Insights Into Pathogenesis.Front Microbiol. 2021 Jan 14;11:614231. doi: 10.3389/fmicb.2020.614231. eCollection 2020. Front Microbiol. 2021. PMID: 33584579 Free PMC article. Review.
-
The Use of Engineered Plant Viruses in a Trans-Kingdom Silencing Strategy Against Their Insect Vectors.Front Plant Sci. 2020 Jul 8;11:917. doi: 10.3389/fpls.2020.00917. eCollection 2020. Front Plant Sci. 2020. PMID: 32733507 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
- 2017YFD0200406, 2016YFC1200603, 2017YFD0200904/National Key Plan for Scientific Research and Development of China
- XDB11040000, XDB11050000/Strategic Priority Research Program of the Chinese Academy of Sciences (CAS)
- 2016ZX08010001/Ministry of Agriculture transgenic major projects
- NSFC 91540116, 31772162, 31472051, 31471782/Natural Science Foundation of China
- Chinese IPM 1801, Chinese IPM 1605/Open Research Fund Program of State Key Laboratory of Integrated Management of Pest Insects and Rodents
LinkOut - more resources
Full Text Sources

