Comparative analysis of rodent and small mammal viromes to better understand the wildlife origin of emerging infectious diseases
- PMID: 30285857
- PMCID: PMC6171170
- DOI: 10.1186/s40168-018-0554-9
Comparative analysis of rodent and small mammal viromes to better understand the wildlife origin of emerging infectious diseases
Abstract
Background: Rodents represent around 43% of all mammalian species, are widely distributed, and are the natural reservoirs of a diverse group of zoonotic viruses, including hantaviruses, Lassa viruses, and tick-borne encephalitis viruses. Thus, analyzing the viral diversity harbored by rodents could assist efforts to predict and reduce the risk of future emergence of zoonotic viral diseases.
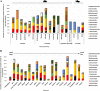
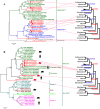
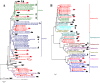
Results: We used next-generation sequencing metagenomic analysis to survey for a range of mammalian viral families in rodents and other small animals of the orders Rodentia, Lagomorpha, and Soricomorpha in China. We sampled 3,055 small animals from 20 provinces and then outlined the spectra of mammalian viruses within these individuals and the basic ecological and genetic characteristics of novel rodent and shrew viruses among the viral spectra. Further analysis revealed that host taxonomy plays a primary role and geographical location plays a secondary role in determining viral diversity. Many viruses were reported for the first time with distinct evolutionary lineages, and viruses related to known human or animal pathogens were identified. Phylogram comparison between viruses and hosts indicated that host shifts commonly happened in many different species during viral evolutionary history.
Conclusions: These results expand our understanding of the viromes of rodents and insectivores in China and suggest that there is high diversity of viruses awaiting discovery in these species in Asia. These findings, combined with our previous bat virome data, greatly increase our knowledge of the viral community in wildlife in a densely populated country in an emerging disease hotspot.
Keywords: Emerging infectious diseases; Rodents; Small mammals; Viral evolution; Virome.
Conflict of interest statement
Ethics approval and consent to participate
Animals were treated according to the guidelines of Regulations for the Administration of Laboratory Animals (Decree No. 2 of the State Science and Technology Commission of the People’s Republic of China, 1988). The sampling procedure was approved by the Ethics Committee of the Institute of Pathogen Biology, Chinese Academy of Medical Sciences & Peking Union Medical College (approval number: IPB EC20100415).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures



Similar articles
-
Decoding the RNA viromes in rodent lungs provides new insight into the origin and evolutionary patterns of rodent-borne pathogens in Mainland Southeast Asia.Microbiome. 2021 Jan 21;9(1):18. doi: 10.1186/s40168-020-00965-z. Microbiome. 2021. PMID: 33478588 Free PMC article.
-
Deciphering the bat virome catalog to better understand the ecological diversity of bat viruses and the bat origin of emerging infectious diseases.ISME J. 2016 Mar;10(3):609-20. doi: 10.1038/ismej.2015.138. Epub 2015 Aug 11. ISME J. 2016. PMID: 26262818 Free PMC article.
-
Assessing the Diversity of Rodent-Borne Viruses: Exploring of High-Throughput Sequencing and Classical Amplification/Sequencing Approaches.Adv Virus Res. 2017;99:61-108. doi: 10.1016/bs.aivir.2017.08.002. Epub 2017 Sep 28. Adv Virus Res. 2017. PMID: 29029730
-
Next generation sequencing technologies: tool to study avian virus diversity.Acta Virol. 2015 Mar;59(1):3-13. doi: 10.4149/av_2015_01_3. Acta Virol. 2015. PMID: 25790045 Review.
-
Global Diversity and Distribution of Hantaviruses and Their Hosts.Ecohealth. 2018 Mar;15(1):163-208. doi: 10.1007/s10393-017-1305-2. Epub 2018 Apr 30. Ecohealth. 2018. PMID: 29713899 Review.
Cited by
-
Identification of a Novel Hepacivirus in Southeast Asian Shrew (Crocidura fuliginosa) from Yunnan Province, China.Pathogens. 2023 Nov 28;12(12):1400. doi: 10.3390/pathogens12121400. Pathogens. 2023. PMID: 38133285 Free PMC article.
-
Abundant and Diverse RNA Viruses in Insects Revealed by RNA-Seq Analysis: Ecological and Evolutionary Implications.mSystems. 2020 Jul 7;5(4):e00039-20. doi: 10.1128/mSystems.00039-20. mSystems. 2020. PMID: 32636338 Free PMC article.
-
Epidemiology and Genomic characteristics of arenavirus in rodents from the southeast coast of P.R. China.BMC Vet Res. 2023 Nov 29;19(1):253. doi: 10.1186/s12917-023-03798-8. BMC Vet Res. 2023. PMID: 38031051 Free PMC article.
-
Murine adenoviruses: tools for studying adenovirus pathogenesis in a natural host.FEBS Lett. 2019 Dec;593(24):3649-3659. doi: 10.1002/1873-3468.13699. Epub 2019 Dec 6. FEBS Lett. 2019. PMID: 31777948 Free PMC article. Review.
-
Identification of Diverse Bat Alphacoronaviruses and Betacoronaviruses in China Provides New Insights Into the Evolution and Origin of Coronavirus-Related Diseases.Front Microbiol. 2019 Aug 14;10:1900. doi: 10.3389/fmicb.2019.01900. eCollection 2019. Front Microbiol. 2019. PMID: 31474969 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

