Role of the Conserved DECH-Box Cysteine in Coupling Hepatitis C Virus Helicase-Catalyzed ATP Hydrolysis to RNA Unwinding
- PMID: 30281972
- PMCID: PMC8532174
- DOI: 10.1021/acs.biochem.8b00796
Role of the Conserved DECH-Box Cysteine in Coupling Hepatitis C Virus Helicase-Catalyzed ATP Hydrolysis to RNA Unwinding
Abstract
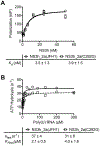
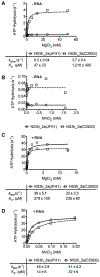
DECH-box proteins are a subset of DExH/D-box superfamily 2 helicases possessing a conserved Asp-Glu-Cys-His motif in their ATP binding site. The conserved His helps position the Asp and Glu residues, which coordinate the divalent metal cation that connects the protein to ATP and activate the water molecule needed for ATP hydrolysis, but the role of the Cys is still unclear. This study uses site-directed mutants of the model DECH-box helicase encoded by the hepatitis C virus (HCV) to examine the role of the Cys in helicase action. Proteins lacking a Cys unwound DNA less efficiently than wild-type proteins did. For example, at low protein concentrations, a helicase harboring a Gly instead of the DECH-box Cys unwound DNA more slowly than the wild-type helicase did, but at higher protein concentrations, the two proteins unwound DNA at similar rates. All HCV proteins analyzed had similar affinities for ATP and nucleic acids and hydrolyzed ATP in the presence of RNA at similar rates. However, in the absence of RNA, all proteins lacking a DECH-box cysteine hydrolyzed ATP 10-15 times faster with higher Km values, and lower apparent affinities for metal ions, compared to those observed with wild-type proteins. These differences were observed with proteins isolated from HCV genotypes 2a and 1b, suggesting that this role is conserved. These data suggest the helicase needs Cys292 to bind ATP in a state where ATP is not hydrolyzed until RNA binds.
Conflict of interest statement
The authors declare no competing financial interest.
Figures



Similar articles
-
Two novel conserved motifs in the hepatitis C virus NS3 protein critical for helicase action.J Biol Chem. 2003 Nov 7;278(45):44514-24. doi: 10.1074/jbc.M306444200. Epub 2003 Aug 27. J Biol Chem. 2003. PMID: 12944414 Free PMC article.
-
Fuel specificity of the hepatitis C virus NS3 helicase.J Mol Biol. 2009 May 15;388(4):851-64. doi: 10.1016/j.jmb.2009.03.059. Epub 2009 Mar 28. J Mol Biol. 2009. PMID: 19332076 Free PMC article.
-
Enhanced nucleic acid binding to ATP-bound hepatitis C virus NS3 helicase at low pH activates RNA unwinding.Nucleic Acids Res. 2004 Aug 2;32(13):4060-70. doi: 10.1093/nar/gkh743. Print 2004. Nucleic Acids Res. 2004. PMID: 15289579 Free PMC article.
-
Primuline derivatives that mimic RNA to stimulate hepatitis C virus NS3 helicase-catalyzed ATP hydrolysis.J Biol Chem. 2013 Jul 5;288(27):19949-57. doi: 10.1074/jbc.M113.463166. Epub 2013 May 23. J Biol Chem. 2013. PMID: 23703611 Free PMC article.
-
Structure and function of hepatitis C virus NS3 helicase.Curr Top Microbiol Immunol. 2000;242:171-96. doi: 10.1007/978-3-642-59605-6_9. Curr Top Microbiol Immunol. 2000. PMID: 10592661 Review.
Cited by
-
The Evolution and Characterization of the RNA Interference Pathways in Lophotrochozoa.Genome Biol Evol. 2024 May 2;16(5):evae098. doi: 10.1093/gbe/evae098. Genome Biol Evol. 2024. PMID: 38713108 Free PMC article.
-
The Discovery of Novel Ferulic Acid Derivatives Incorporating Substituted Isopropanolamine Moieties as Potential Tobacco Mosaic Virus Helicase Inhibitors.Int J Mol Sci. 2022 Nov 13;23(22):13991. doi: 10.3390/ijms232213991. Int J Mol Sci. 2022. PMID: 36430473 Free PMC article.
References
-
- Choo QL, Kuo G, Weiner AJ, Overby LR, Bradley DW, and Houghton M (1989) Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science 244, 359–362. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

