Structure of glyoxysomal malate dehydrogenase (MDH3) from Saccharomyces cerevisiae
- PMID: 30279312
- PMCID: PMC6168765
- DOI: 10.1107/S2053230X18011895
Structure of glyoxysomal malate dehydrogenase (MDH3) from Saccharomyces cerevisiae
Abstract
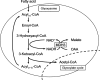
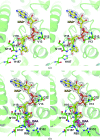
Malate dehydrogenase (MDH), a carbohydrate and energy metabolism enzyme in eukaryotes, catalyzes the interconversion of malate to oxaloacetate (OAA) in conjunction with that of nicotinamide adenine dinucleotide (NAD+) to NADH. Three isozymes of MDH have been reported in Saccharomyces cerevisiae: MDH1, MDH2 and MDH3. MDH1 is a mitochondrial enzyme and a member of the tricarboxylic acid cycle, whereas MDH2 is a cytosolic enzyme that functions in the glyoxylate cycle. MDH3 is a glyoxysomal enzyme that is involved in the reoxidation of NADH, which is produced during fatty-acid β-oxidation. The affinity of MDH3 for OAA is lower than those of MDH1 and MDH2. Here, the crystal structures of yeast apo MDH3, the MDH3-NAD+ complex and the MDH3-NAD+-OAA ternary complex were determined. The structure of the ternary complex suggests that the active-site loop is in the open conformation, differing from the closed conformations in mitochondrial and cytosolic malate dehydrogenases.
Keywords: MDH3; Saccharomyces cerevisiae; X-ray crystallography; fatty-acid β-oxidation; glyoxysome; malate dehydrogenase.
Figures

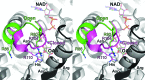
Similar articles
-
Conformational changes on substrate binding revealed by structures of Methylobacterium extorquens malate dehydrogenase.Acta Crystallogr F Struct Biol Commun. 2018 Oct 1;74(Pt 10):610-616. doi: 10.1107/S2053230X18011809. Epub 2018 Sep 19. Acta Crystallogr F Struct Biol Commun. 2018. PMID: 30279311 Free PMC article.
-
Isolation and characterization of the yeast gene encoding the MDH3 isozyme of malate dehydrogenase.J Biol Chem. 1992 Dec 5;267(34):24708-15. J Biol Chem. 1992. PMID: 1447211
-
High-resolution crystal structure of the reduced Grx1 from Saccharomyces cerevisiae.Acta Crystallogr F Struct Biol Commun. 2019 May 1;75(Pt 5):392-396. doi: 10.1107/S2053230X19003327. Epub 2019 Apr 26. Acta Crystallogr F Struct Biol Commun. 2019. PMID: 31045569 Free PMC article.
-
Expression and function of a mislocalized form of peroxisomal malate dehydrogenase (MDH3) in yeast.J Biol Chem. 1995 Sep 8;270(36):21220-5. doi: 10.1074/jbc.270.36.21220. J Biol Chem. 1995. PMID: 7673155
-
Malate dehydrogenases--structure and function.Gen Physiol Biophys. 2002 Sep;21(3):257-65. Gen Physiol Biophys. 2002. PMID: 12537350 Review.
Cited by
-
Antifungal activity of 2-chloro-5-trifluoromethoxybenzeneboronic acid and inhibitory mechanisms on Geotrichum candidum from sour rot Xiaozhou mustard root tuber.Sci Rep. 2024 Oct 1;14(1):22802. doi: 10.1038/s41598-024-74211-z. Sci Rep. 2024. PMID: 39354024 Free PMC article.
-
Identification and characterization of cytosolic malate dehydrogenase from the liver fluke Fasciola gigantica.Sci Rep. 2020 Aug 7;10(1):13372. doi: 10.1038/s41598-020-70202-y. Sci Rep. 2020. PMID: 32770017 Free PMC article.
-
A fragment-based approach identifies an allosteric pocket that impacts malate dehydrogenase activity.Commun Biol. 2021 Aug 10;4(1):949. doi: 10.1038/s42003-021-02442-1. Commun Biol. 2021. PMID: 34376783 Free PMC article.
-
Dynamics of nutrients, sensory quality and microbial communities and their interactions during co-fermentation of pineapple by-products and whey protein.Food Chem X. 2024 Feb 28;22:101254. doi: 10.1016/j.fochx.2024.101254. eCollection 2024 Jun 30. Food Chem X. 2024. PMID: 38444559 Free PMC article.
-
Improved energy efficiency in microbial fuel cells by bioethanol and electricity co-generation.Biotechnol Biofuels Bioprod. 2022 Aug 17;15(1):84. doi: 10.1186/s13068-022-02180-4. Biotechnol Biofuels Bioprod. 2022. PMID: 35978352 Free PMC article.
References
-
- Bell, J. K., Yennawar, H. P., Wright, S. K., Thompson, J. R., Viola, R. E. & Banaszak, L. J. (2001). J. Biol. Chem. 276, 31156–31162. - PubMed
-
- Birktoft, J. J., Rhodes, G. & Banaszak, L. J. (1989). Biochemistry, 28, 6065–6081. - PubMed
-
- Clarke, A. R., Wigley, D. B., Chia, W. N., Barstow, D., Atkinson, T. & Holbrook, J. J. (1986). Nature (London), 324, 699–702. - PubMed
-
- Cox, B., Chit, M. M., Weaver, T., Gietl, C., Bailey, J., Bell, E. & Banaszak, L. (2005). FEBS J. 272, 643–654. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

