Atrial Structural Remodeling Gene Variants in Patients with Atrial Fibrillation
- PMID: 30276209
- PMCID: PMC6151856
- DOI: 10.1155/2018/4862480
Atrial Structural Remodeling Gene Variants in Patients with Atrial Fibrillation
Abstract
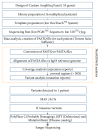
Atrial fibrillation (AF) is a common arrhythmia for which the genetic studies mainly focused on the genes involved in electrical remodeling, rather than left atrial muscle remodeling. To identify rare variants involved in atrial myopathy using mutational screening, a high-throughput next-generation sequencing (NGS) workflow was developed based on a custom AmpliSeq™ panel of 55 genes potentially involved in atrial myopathy. This workflow was applied to a cohort of 94 patients with AF, 76 with atrial dilatation and 18 without. Bioinformatic analyses used NextGENe® software and in silico tools for variant interpretation. The AmpliSeq custom-made panel efficiently explored 96.58% of the targeted sequences. Based on in silico analysis, 11 potentially pathogenic missense variants were identified that were not previously associated with AF. These variants were located in genes involved in atrial tissue structural remodeling. Three patients were also carriers of potential variants in prevalent arrhythmia-causing genes, usually associated with AF. Most of the variants were found in patients with atrial dilatation (n=9, 82%). This NGS approach was a sensitive and specific method that identified 11 potentially pathogenic variants, which are likely to play roles in the predisposition to left atrial myopathy. Functional studies are needed to confirm their pathogenicity.
Figures


Similar articles
-
Deep sequencing of atrial fibrillation patients with mitral valve regurgitation shows no evidence of mosaicism but reveals novel rare germline variants.Heart Rhythm. 2017 Oct;14(10):1531-1538. doi: 10.1016/j.hrthm.2017.05.027. Epub 2017 May 24. Heart Rhythm. 2017. PMID: 28549997
-
Targeted deep sequencing reveals no definitive evidence for somatic mosaicism in atrial fibrillation.Circ Cardiovasc Genet. 2015 Feb;8(1):50-7. doi: 10.1161/CIRCGENETICS.114.000650. Epub 2014 Nov 18. Circ Cardiovasc Genet. 2015. PMID: 25406240 Free PMC article. Clinical Trial.
-
Post-operative atrial fibrillation examined using whole-genome RNA sequencing in human left atrial tissue.BMC Med Genomics. 2017 May 2;10(1):25. doi: 10.1186/s12920-017-0270-5. BMC Med Genomics. 2017. PMID: 28464817 Free PMC article. Clinical Trial.
-
Is There a Role for Genes in Exercise-Induced Atrial Cardiomyopathy?Heart Lung Circ. 2018 Sep;27(9):1093-1098. doi: 10.1016/j.hlc.2018.03.028. Epub 2018 Apr 9. Heart Lung Circ. 2018. PMID: 29706494 Review.
-
Atrial fibrillation from the pathologist's perspective.Cardiovasc Pathol. 2014 Mar-Apr;23(2):71-84. doi: 10.1016/j.carpath.2013.12.001. Epub 2013 Dec 15. Cardiovasc Pathol. 2014. PMID: 24462196 Review.
Cited by
-
One gene, two modes of inheritance, four diseases: A systematic review of the cardiac manifestation of pathogenic variants in JPH2-encoded junctophilin-2.Trends Cardiovasc Med. 2023 Jan;33(1):1-10. doi: 10.1016/j.tcm.2021.11.006. Epub 2021 Dec 1. Trends Cardiovasc Med. 2023. PMID: 34861382 Free PMC article. Review.
-
Epigenetics in atrial fibrillation: A reappraisal.Heart Rhythm. 2021 May;18(5):824-832. doi: 10.1016/j.hrthm.2021.01.007. Epub 2021 Jan 10. Heart Rhythm. 2021. PMID: 33440248 Free PMC article. Review.
-
The Effects of Fibrotic Cell Type and Its Density on Atrial Fibrillation Dynamics: An In Silico Study.Cells. 2021 Oct 15;10(10):2769. doi: 10.3390/cells10102769. Cells. 2021. PMID: 34685750 Free PMC article.
-
Genetic Discovery of ATP-Sensitive K+ Channels in Cardiovascular Diseases.Circ Arrhythm Electrophysiol. 2019 May;12(5):e007322. doi: 10.1161/CIRCEP.119.007322. Circ Arrhythm Electrophysiol. 2019. PMID: 31030551 Free PMC article. Review.
-
Left Atrial Cardiomyopathy - A Challenging Diagnosis.Front Cardiovasc Med. 2022 Jun 30;9:942385. doi: 10.3389/fcvm.2022.942385. eCollection 2022. Front Cardiovasc Med. 2022. PMID: 35845077 Free PMC article. Review.
References
-
- Kirchhof P., Lip G. Y. H., Van Gelder I. C., et al. Comprehensive risk reduction in patients with atrial fibrillation: Emerging diagnostic and therapeutic options—a report from the 3rd Atrial Fibrillation Competence NETwork/European Heart Rhythm Association consensus conference. Europace. 2012;14(1):8–27. doi: 10.1093/europace/eur241. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

