Restriction-deficient mutants and marker-less genomic modification for metabolic engineering of the solvent producer Clostridium saccharobutylicum
- PMID: 30275904
- PMCID: PMC6158908
- DOI: 10.1186/s13068-018-1260-3
Restriction-deficient mutants and marker-less genomic modification for metabolic engineering of the solvent producer Clostridium saccharobutylicum
Abstract
Background: Clostridium saccharobutylicum NCP 262 is a solventogenic bacterium that has been used for the industrial production of acetone, butanol, and ethanol. The lack of a genetic manipulation system for C. saccharobutylicum currently limits (i) the use of metabolic pathway engineering to improve the yield, titer, and productivity of n-butanol production by this microorganism, and (ii) functional genomics studies to better understand its physiology.
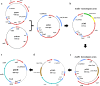
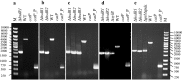
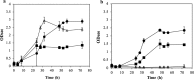
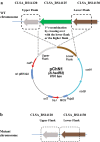
Results: In this study, a marker-less deletion system was developed for C. saccharobutylicum using the codBA operon genes from Clostridium ljungdahlii as a counterselection marker. The codB gene encodes a cytosine permease, while codA encodes a cytosine deaminase that converts 5-fluorocytosine to 5-fluorouracil, which is toxic to the cell. To introduce a marker-less genomic modification, we constructed a suicide vector containing: the catP gene for thiamphenicol resistance; the codBA operon genes for counterselection; fused DNA segments both upstream and downstream of the chromosomal deletion target. This vector was introduced into C. saccharobutylicum by tri-parental conjugation. Single crossover integrants are selected on plates supplemented with thiamphenicol and colistin, and, subsequently, double-crossover mutants whose targeted chromosomal sequence has been deleted were identified by counterselection on plates containing 5-fluorocytosine. Using this marker-less deletion system, we constructed the restriction-deficient mutant C. saccharobutylicum ΔhsdR1ΔhsdR2ΔhsdR3, which we named C. saccharobutylicum Ch2. This triple mutant exhibits high transformation efficiency with unmethylated DNA. To demonstrate its applicability to metabolic engineering, the method was first used to delete the xylB gene to study its role in xylose and arabinose metabolism. Furthermore, we also deleted the ptb and buk genes to create a butyrate metabolism-negative mutant of C. saccharobutylicum that produces n-butanol at high yield.
Conclusions: The plasmid vectors and the method introduced here, together with the restriction-deficient strains described in this work, for the first time, allow for efficient marker-less genomic modification of C. saccharobutylicum and, therefore, represent valuable tools for the genetic and metabolic engineering of this industrially important solvent-producing organism.
Keywords: 5-Fluorocytosine; Butyrate kinase; CodB/codA; Phosphotransbutyrylase conjugation; Xylulose kinase.
Figures
Similar articles
-
An efficient method for markerless mutant generation by allelic exchange in Clostridium acetobutylicum and Clostridium saccharobutylicum using suicide vectors.Biotechnol Biofuels. 2019 Feb 14;12:31. doi: 10.1186/s13068-019-1364-4. eCollection 2019. Biotechnol Biofuels. 2019. PMID: 30809274 Free PMC article.
-
Engineering coenzyme A-dependent pathway from Clostridium saccharobutylicum in Escherichia coli for butanol production.Bioresour Technol. 2017 Jul;235:140-148. doi: 10.1016/j.biortech.2017.03.085. Epub 2017 Mar 18. Bioresour Technol. 2017. PMID: 28365341
-
Construction of a restriction-less, marker-less mutant useful for functional genomic and metabolic engineering of the biofuel producer Clostridium acetobutylicum.Biotechnol Biofuels. 2016 Feb 2;9:23. doi: 10.1186/s13068-016-0432-2. eCollection 2016. Biotechnol Biofuels. 2016. PMID: 26839586 Free PMC article.
-
Recent advances in n-butanol and butyrate production using engineered Clostridium tyrobutyricum.World J Microbiol Biotechnol. 2020 Aug 14;36(9):138. doi: 10.1007/s11274-020-02914-2. World J Microbiol Biotechnol. 2020. PMID: 32794091 Review.
-
Engineering Clostridium for improved solvent production: recent progress and perspective.Appl Microbiol Biotechnol. 2019 Jul;103(14):5549-5566. doi: 10.1007/s00253-019-09916-7. Epub 2019 May 29. Appl Microbiol Biotechnol. 2019. PMID: 31139901 Review.
Cited by
-
The Roles of the Various Cellulose Biosynthesis Operons in Komagataeibacter hansenii ATCC 23769.Appl Environ Microbiol. 2022 Apr 12;88(7):e0246021. doi: 10.1128/aem.02460-21. Epub 2022 Mar 23. Appl Environ Microbiol. 2022. PMID: 35319232 Free PMC article.
-
Role of butyrogenic Firmicutes in type-2 diabetes.J Diabetes Metab Disord. 2022 Jul 14;21(2):1873-1882. doi: 10.1007/s40200-022-01081-5. eCollection 2022 Dec. J Diabetes Metab Disord. 2022. PMID: 36404833 Free PMC article. Review.
-
Analysis of cellulose synthesis in a high-producing acetic acid bacterium Komagataeibacter hansenii.Appl Microbiol Biotechnol. 2023 May;107(9):2947-2967. doi: 10.1007/s00253-023-12461-z. Epub 2023 Mar 17. Appl Microbiol Biotechnol. 2023. PMID: 36930278 Free PMC article.
-
An efficient method for markerless mutant generation by allelic exchange in Clostridium acetobutylicum and Clostridium saccharobutylicum using suicide vectors.Biotechnol Biofuels. 2019 Feb 14;12:31. doi: 10.1186/s13068-019-1364-4. eCollection 2019. Biotechnol Biofuels. 2019. PMID: 30809274 Free PMC article.
-
The Restriction-Modification Systems of Clostridium carboxidivorans P7.Microorganisms. 2023 Dec 12;11(12):2962. doi: 10.3390/microorganisms11122962. Microorganisms. 2023. PMID: 38138106 Free PMC article.
References
LinkOut - more resources
Full Text Sources

