Exploration of Survival Traits, Probiotic Determinants, Host Interactions, and Functional Evolution of Bifidobacterial Genomes Using Comparative Genomics
- PMID: 30275399
- PMCID: PMC6210967
- DOI: 10.3390/genes9100477
Exploration of Survival Traits, Probiotic Determinants, Host Interactions, and Functional Evolution of Bifidobacterial Genomes Using Comparative Genomics
Abstract
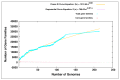
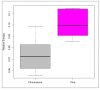
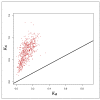
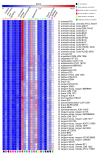
Members of the genus Bifidobacterium are found in a wide-range of habitats and are used as important probiotics. Thus, exploration of their functional traits at the genus level is of utmost significance. Besides, this genus has been demonstrated to exhibit an open pan-genome based on the limited number of genomes used in earlier studies. However, the number of genomes is a crucial factor for pan-genome calculations. We have analyzed the pan-genome of a comparatively larger dataset of 215 members of the genus Bifidobacterium belonging to different habitats, which revealed an open nature. The pan-genome for the 56 probiotic and human-gut strains of this genus, was also found to be open. The accessory- and unique-components of this pan-genome were found to be under the operation of Darwinian selection pressure. Further, their genome-size variation was predicted to be attributed to the abundance of certain functions carried by genomic islands, which are facilitated by insertion elements and prophages. In silico functional and host-microbe interaction analyses of their core-genome revealed significant genomic factors for niche-specific adaptations and probiotic traits. The core survival traits include stress tolerance, biofilm formation, nutrient transport, and Sec-secretion system, whereas the core probiotic traits are imparted by the factors involved in carbohydrate- and protein-metabolism and host-immunomodulations.
Keywords: Bifidobacterium; comparative genomics; host-microbe interaction; human-gut; immunomodulations; niche-specific adaptations; pan-genome; probiotic.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Comparative analysis of probiotic bacteria based on a new definition of core genome.J Bioinform Comput Biol. 2018 Jun;16(3):1840012. doi: 10.1142/S0219720018400127. Epub 2018 Mar 26. J Bioinform Comput Biol. 2018. PMID: 29792377
-
Pan-genome evolution and its association with divergence of metabolic functions in Bifidobacterium genus.World J Microbiol Biotechnol. 2022 Oct 7;38(12):231. doi: 10.1007/s11274-022-03430-1. World J Microbiol Biotechnol. 2022. PMID: 36205822
-
Comparative genomics of Bifidobacterium, Lactobacillus and related probiotic genera.Microb Ecol. 2012 Apr;63(3):651-73. doi: 10.1007/s00248-011-9948-y. Epub 2011 Oct 27. Microb Ecol. 2012. PMID: 22031452 Free PMC article. Review.
-
Pan-genome analysis of six Paracoccus type strain genomes reveal lifestyle traits.PLoS One. 2023 Dec 20;18(12):e0287947. doi: 10.1371/journal.pone.0287947. eCollection 2023. PLoS One. 2023. PMID: 38117845 Free PMC article.
-
Probiotic characters of Bifidobacterium and Lactobacillus are a result of the ongoing gene acquisition and genome minimization evolutionary trends.Microb Pathog. 2017 Oct;111:118-131. doi: 10.1016/j.micpath.2017.08.021. Epub 2017 Aug 18. Microb Pathog. 2017. PMID: 28826768 Review.
Cited by
-
Complete Genome Sequencing and Comparative Genomics of Three Potential Probiotic Strains, Lacticaseibacillus casei FBL6, Lacticaseibacillus chiayiensis FBL7, and Lacticaseibacillus zeae FBL8.Front Microbiol. 2022 Jan 7;12:794315. doi: 10.3389/fmicb.2021.794315. eCollection 2021. Front Microbiol. 2022. PMID: 35069490 Free PMC article.
-
Bifidobacterium β-Glucosidase Activity and Fermentation of Dietary Plant Glucosides Is Species and Strain Specific.Microorganisms. 2020 Jun 3;8(6):839. doi: 10.3390/microorganisms8060839. Microorganisms. 2020. PMID: 32503148 Free PMC article.
-
Evolutionary relationships among bifidobacteria and their hosts and environments.BMC Genomics. 2020 Jan 8;21(1):26. doi: 10.1186/s12864-019-6435-1. BMC Genomics. 2020. PMID: 31914919 Free PMC article.
-
Enterotype Variations of the Healthy Human Gut Microbiome in Different Geographical Regions.Bioinformation. 2018 Dec 29;14(9):560-573. doi: 10.6026/97320630014560. eCollection 2018. Bioinformation. 2018. PMID: 31223215 Free PMC article.
-
Comparative gut microbiome analysis of the Prakriti and Sasang systems reveals functional level similarities in constitutionally similar classes.3 Biotech. 2020 Sep;10(9):379. doi: 10.1007/s13205-020-02376-1. Epub 2020 Aug 7. 3 Biotech. 2020. PMID: 32802721 Free PMC article.
References
-
- Pinto-Sánchez M.I., Smecuol E.C., Temprano M.P., Sugai E., González A., Moreno M.L., Huang X., Bercik P., Cabanne A., Vázquez H. Bifidobacterium infantis NLS super strain reduces the expression of α-defensin-5, a marker of innate immunity, in the mucosa of active celiac disease patients. J. Clin. Gastroenterol. 2017;51:814–817. doi: 10.1097/MCG.0000000000000687. - DOI - PubMed
-
- Sánchez B., Champomier-Verges M.-C., del Carmen Collado M., Anglade P., Baraige F., Sanz Y., Clara G., Margolles A., Zagorec M. Low-pH adaptation and the acid tolerance response of Bifidobacterium longum biotype longum. Appl. Environ. Microbiol. 2007;73:6450–6459. doi: 10.1128/AEM.00886-07. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

