NKX2-1-AS1 negatively regulates CD274/PD-L1, cell-cell interaction genes, and limits human lung carcinoma cell migration
- PMID: 30258080
- PMCID: PMC6158174
- DOI: 10.1038/s41598-018-32793-5
NKX2-1-AS1 negatively regulates CD274/PD-L1, cell-cell interaction genes, and limits human lung carcinoma cell migration
Abstract
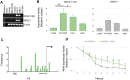
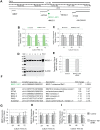
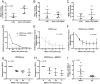
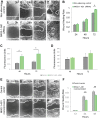
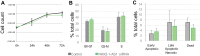
The function of most long noncoding RNAs (lncRNAs) is unknown. However, recent studies reveal important roles of lncRNAs in regulating cancer-related pathways. Human antisense lncRNA-NKX2-1-AS1 partially overlaps the NKX2-1/TTF1 gene within chromosomal region 14q13.3. Amplification of this region and/or differential expression of genes therein are associated with cancer progression. Herein we show higher levels of NKX2-AS1 and NKX2-1 in lung adenocarcinomas relative to non-tumor controls but no correlation between NKX2-1-AS1 and NKX2-1 levels across specimens, or with amplification of the 14q13.3 region, suggesting that NKX2-1-AS1 and NKX2-1 are independently regulated. Loss-and-gain of function experiments showed that NKX2-1-AS1 does not regulate NKX2-1 expression, or nearby genes, but controls genes in trans. Genes up-regulated by NKX2-1-AS1-knockdown belong to cell adhesion and PD-L1/PD-1 checkpoint pathways. NKX2-1-AS1 negatively regulates endogenous CD274/PD-L1, a known target of NKX2-1, and the transcriptional activity of -1kb-CD274 promoter-reporter construct. Furthermore, NKX2-1-AS1 interferes with NKX2-1 protein binding to the CD274-promoter, likely by NKX2-1 protein-NKX2-1-AS1 interactions. Finally, NKX2-1-AS1 negatively regulates cell migration and wound healing, but not proliferation or apoptosis. These findings support potential roles of NKX2-1-AS1 in limiting motility and immune system evasion of lung carcinoma cells, highlighting a novel mechanism that may influence tumorigenic capabilities of lung epithelial cells.
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
A natural antisense lncRNA controls breast cancer progression by promoting tumor suppressor gene mRNA stability.PLoS Genet. 2018 Nov 29;14(11):e1007802. doi: 10.1371/journal.pgen.1007802. eCollection 2018 Nov. PLoS Genet. 2018. PMID: 30496290 Free PMC article.
-
Targeting the lncRNA FGD5-AS1/miR-497-5p/PD-L1 Axis Inhibits Malignant Phenotypes in Colon Cancer (CC).Biomed Res Int. 2022 Jul 6;2022:1133332. doi: 10.1155/2022/1133332. eCollection 2022. Biomed Res Int. 2022. Retraction in: Biomed Res Int. 2023 Oct 4;2023:9864810. doi: 10.1155/2023/9864810 PMID: 35845947 Free PMC article. Retracted.
-
LncRNA FGD5-AS1 promotes the malignant phenotypes of ovarian cancer cells via targeting miR-142-5p.Apoptosis. 2021 Jun;26(5-6):348-360. doi: 10.1007/s10495-021-01674-0. Epub 2021 May 11. Apoptosis. 2021. PMID: 33974163
-
YY1-induced long non-coding RNA PSMA3 antisense RNA 1 functions as a competing endogenous RNA for microRNA 214-5p to expedite the viability and restrict the apoptosis of bladder cancer cells via regulating programmed cell death-ligand 1.Bioengineered. 2021 Dec;12(2):9150-9161. doi: 10.1080/21655979.2021.1994907. Bioengineered. 2021. PMID: 34720049 Free PMC article.
-
AFAP1-AS1: a rising star among oncogenic long non-coding RNAs.Sci China Life Sci. 2021 Oct;64(10):1602-1611. doi: 10.1007/s11427-020-1874-6. Epub 2021 May 13. Sci China Life Sci. 2021. PMID: 33999309 Review.
Cited by
-
Focus on PD-1/PD-L1 as a Therapeutic Target in Ovarian Cancer.Int J Mol Sci. 2022 Oct 11;23(20):12067. doi: 10.3390/ijms232012067. Int J Mol Sci. 2022. PMID: 36292922 Free PMC article. Review.
-
An immunotherapeutic approach to decipher the role of long non-coding RNAs in cancer progression, resistance and epigenetic regulation of immune cells.J Exp Clin Cancer Res. 2021 Jul 24;40(1):242. doi: 10.1186/s13046-021-01997-5. J Exp Clin Cancer Res. 2021. PMID: 34303380 Free PMC article. Review.
-
Soluble PD-1: Predictive, Prognostic, and Therapeutic Value for Cancer Immunotherapy.Front Immunol. 2020 Nov 19;11:587460. doi: 10.3389/fimmu.2020.587460. eCollection 2020. Front Immunol. 2020. PMID: 33329567 Free PMC article. Review.
-
LncRNA CBR3-AS1 potentiates Wnt/β-catenin signaling to regulate lung adenocarcinoma cells proliferation, migration and invasion.Cancer Cell Int. 2021 Jan 9;21(1):36. doi: 10.1186/s12935-020-01685-y. Cancer Cell Int. 2021. PMID: 33422081 Free PMC article.
-
OTUD6B-AS1 Might Be a Novel Regulator of Apoptosis in Systemic Sclerosis.Front Immunol. 2019 May 17;10:1100. doi: 10.3389/fimmu.2019.01100. eCollection 2019. Front Immunol. 2019. PMID: 31156645 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

