Proteomic analysis of rat serum revealed the effects of chronic sleep deprivation on metabolic, cardiovascular and nervous system
- PMID: 30235220
- PMCID: PMC6147403
- DOI: 10.1371/journal.pone.0199237
Proteomic analysis of rat serum revealed the effects of chronic sleep deprivation on metabolic, cardiovascular and nervous system
Abstract
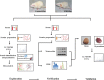
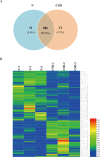
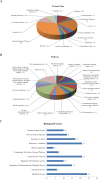
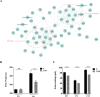
Sleep is an essential and fundamental physiological process that plays crucial roles in the balance of psychological and physical health. Sleep disorder may lead to adverse health outcomes. The effects of sleep deprivation were extensively studied, but its mechanism is still not fully understood. The present study aimed to identify the alterations of serum proteins associated with chronic sleep deprivation, and to seek for potential biomarkers of sleep disorder mediated diseases. A label-free quantitative proteomics technology was used to survey the global changes of serum proteins between normal rats and chronic sleep deprivation rats. A total of 309 proteins were detected in the serum samples and among them, 117 proteins showed more than 1.8-folds abundance alterations between the two groups. Functional enrichment and network analyses of the differential proteins revealed a close relationship between chronic sleep deprivation and several biological processes including energy metabolism, cardiovascular function and nervous function. And four proteins including pyruvate kinase M1, clusterin, kininogen1 and profilin-1were identified as potential biomarkers for chronic sleep deprivation. The four candidates were validated via parallel reaction monitoring (PRM) based targeted proteomics. In addition, protein expression alteration of the four proteins was confirmed in myocardium and brain of rat model. In summary, the comprehensive proteomic study revealed the biological impacts of chronic sleep deprivation and discovered several potential biomarkers. This study provides further insight into the pathological and molecular mechanisms underlying sleep disorders at protein level.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Similar articles
-
The short- and long-term proteomic effects of sleep deprivation on the cortical and thalamic synapses.Mol Cell Neurosci. 2017 Mar;79:64-80. doi: 10.1016/j.mcn.2017.01.002. Epub 2017 Jan 10. Mol Cell Neurosci. 2017. PMID: 28087334
-
Integrated metabolomics and proteomics analysis reveals energy metabolism disorders in the livers of sleep-deprived mice.J Proteomics. 2021 Aug 15;245:104290. doi: 10.1016/j.jprot.2021.104290. Epub 2021 Jun 2. J Proteomics. 2021. PMID: 34089895
-
The Effect of Sleep Deprivation and Subsequent Recovery Period on the Synaptic Proteome of Rat Cerebral Cortex.Mol Neurobiol. 2022 Feb;59(2):1301-1319. doi: 10.1007/s12035-021-02699-x. Epub 2022 Jan 5. Mol Neurobiol. 2022. PMID: 34988919 Free PMC article.
-
Functional consequences of sustained sleep deprivation in the rat.Behav Brain Res. 1995 Jul-Aug;69(1-2):43-54. doi: 10.1016/0166-4328(95)00009-i. Behav Brain Res. 1995. PMID: 7546317 Review.
-
Cellular consequences of sleep deprivation in the brain.Sleep Med Rev. 2006 Oct;10(5):307-21. doi: 10.1016/j.smrv.2006.04.001. Epub 2006 Aug 21. Sleep Med Rev. 2006. PMID: 16920372 Review.
Cited by
-
Neuroprotective effect of Astragali Radix on cerebral infarction based on proteomics.Front Pharmacol. 2023 Jun 9;14:1162134. doi: 10.3389/fphar.2023.1162134. eCollection 2023. Front Pharmacol. 2023. PMID: 37361203 Free PMC article.
-
Chronic Partial Sleep Deprivation Increased the Incidence of Atrial Fibrillation by Promoting Pulmonary Vein and Atrial Arrhythmogenesis in a Rodent Model.Int J Mol Sci. 2024 Jul 11;25(14):7619. doi: 10.3390/ijms25147619. Int J Mol Sci. 2024. PMID: 39062858 Free PMC article.
-
Translational and Posttranslational Dynamics in a Model Peptidergic System.Mol Cell Proteomics. 2023 May;22(5):100544. doi: 10.1016/j.mcpro.2023.100544. Epub 2023 Apr 6. Mol Cell Proteomics. 2023. PMID: 37030596 Free PMC article.
-
Lithium Treatment Improves Cardiac Dysfunction in Rats Deprived of Rapid Eye Movement Sleep.Int J Mol Sci. 2022 Sep 23;23(19):11226. doi: 10.3390/ijms231911226. Int J Mol Sci. 2022. PMID: 36232526 Free PMC article.
-
Proteomics analysis of coronary atherosclerotic heart disease with different Traditional Chinese Medicine syndrome types before and after percutaneous coronary intervention.J Tradit Chin Med. 2024 Jun;44(3):554-563. doi: 10.19852/j.cnki.jtcm.20240408.001. J Tradit Chin Med. 2024. PMID: 38767640 Free PMC article.
References
-
- Brianza-Padilla M, Bonilla-Jaime H, Almanza-Perez JC, Lopez-Lopez AL, Sanchez-Munoz F, Vazquez-Palacios G: Effects of different periods of paradoxical sleep deprivation and sleep recovery on lipid and glucose metabolism and appetite hormones in rats. Appl Physiol Nutr Metab. 2016, 41:235–243. 10.1139/apnm-2015-0337 - DOI - PubMed
-
- Michael H. Bonnet DLA: We are chronically sleep deprived. sleep. 1995, 18:4. - PubMed
-
- Yuan R, Wang J, Guo L-l: The Effect of Sleep Deprivation on Coronary Heart Disease. Chinese Medical Sciences Journal. 2016, 31:247–253. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

