Systematic identification of cancer-specific MHC-binding peptides with RAVEN
- PMID: 30228952
- PMCID: PMC6140548
- DOI: 10.1080/2162402X.2018.1481558
Systematic identification of cancer-specific MHC-binding peptides with RAVEN
Abstract
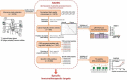
Immunotherapy can revolutionize anti-cancer therapy if specific targets are available. Immunogenic peptides encoded by cancer-specific genes (CSGs) may enable targeted immunotherapy, even of oligo-mutated cancers, which lack neo-antigens generated by protein-coding missense mutations. Here, we describe an algorithm and user-friendly software named RAVEN (Rich Analysis of Variable gene Expressions in Numerous tissues) that automatizes the systematic and fast identification of CSG-encoded peptides highly affine to Major Histocompatibility Complexes (MHC) starting from transcriptome data. We applied RAVEN to a dataset assembled from 2,678 simultaneously normalized gene expression microarrays comprising 50 tumor entities, with a focus on oligo-mutated pediatric cancers, and 71 normal tissue types. RAVEN performed a transcriptome-wide scan in each cancer entity for gender-specific CSGs, and identified several established CSGs, but also many novel candidates potentially suitable for targeting multiple cancer types. The specific expression of the most promising CSGs was validated in cancer cell lines and in a comprehensive tissue-microarray. Subsequently, RAVEN identified likely immunogenic CSG-encoded peptides by predicting their affinity to MHCs and excluded sequence identity to abundantly expressed proteins by interrogating the UniProt protein-database. The predicted affinity of selected peptides was validated in T2-cell peptide-binding assays in which many showed binding-kinetics like a very immunogenic influenza control peptide. Collectively, we provide an exquisitely curated catalogue of cancer-specific and highly MHC-affine peptides across 50 cancer types, and a freely available software (https://github.com/JSGerke/RAVENsoftware) to easily apply our algorithm to any gene expression dataset. We anticipate that our peptide libraries and software constitute a rich resource to advance anti-cancer immunotherapy.
Keywords: Immunotherapy; bioinformatics; cancer-specific genes; microarray.
Figures



Similar articles
-
An integrated database of experimentally validated major histocompatibility complex epitopes for antigen-specific cancer therapy.Antib Ther. 2024 Jun 17;7(2):177-186. doi: 10.1093/abt/tbae011. eCollection 2024 Apr. Antib Ther. 2024. PMID: 38933532 Free PMC article.
-
POPI: predicting immunogenicity of MHC class I binding peptides by mining informative physicochemical properties.Bioinformatics. 2007 Apr 15;23(8):942-9. doi: 10.1093/bioinformatics/btm061. Epub 2007 Mar 24. Bioinformatics. 2007. PMID: 17384427
-
Mutant MHC class II epitopes drive therapeutic immune responses to cancer.Nature. 2015 Apr 30;520(7549):692-6. doi: 10.1038/nature14426. Epub 2015 Apr 22. Nature. 2015. PMID: 25901682 Free PMC article.
-
Candidate epitope identification using peptide property models: application to cancer immunotherapy.Methods. 2004 Dec;34(4):460-7. doi: 10.1016/j.ymeth.2004.06.001. Methods. 2004. PMID: 15542372 Review.
-
Prospects for T cell immunotherapy of tumours by vaccination with immunodominant and subdominant peptides.Ciba Found Symp. 1994;187:97-104; discussion 104-12. doi: 10.1002/9780470514672.ch7. Ciba Found Symp. 1994. PMID: 7796678 Review.
Cited by
-
Glycoproteogenomics: Setting the Course for Next-generation Cancer Neoantigen Discovery for Cancer Vaccines.Genomics Proteomics Bioinformatics. 2021 Feb;19(1):25-43. doi: 10.1016/j.gpb.2021.03.005. Epub 2021 Jun 9. Genomics Proteomics Bioinformatics. 2021. PMID: 34118464 Free PMC article. Review.
-
The Value of Microbes in Cancer Neoantigen Immunotherapy.Pharmaceutics. 2023 Aug 14;15(8):2138. doi: 10.3390/pharmaceutics15082138. Pharmaceutics. 2023. PMID: 37631352 Free PMC article. Review.
-
Targeting the CALCB/RAMP1 axis inhibits growth of Ewing sarcoma.Cell Death Dis. 2019 Feb 11;10(2):116. doi: 10.1038/s41419-019-1372-0. Cell Death Dis. 2019. PMID: 30741933 Free PMC article.
-
Oncogenic hijacking of a developmental transcription factor evokes vulnerability toward oxidative stress in Ewing sarcoma.Nat Commun. 2020 May 15;11(1):2423. doi: 10.1038/s41467-020-16244-2. Nat Commun. 2020. PMID: 32415069 Free PMC article.
-
Current and Future Treatment Strategies for Rhabdomyosarcoma.Front Oncol. 2019 Dec 20;9:1458. doi: 10.3389/fonc.2019.01458. eCollection 2019. Front Oncol. 2019. PMID: 31921698 Free PMC article. Review.
References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
