Losses of cytokines and chemokines are common genetic features of human cancers: the somatic copy number alterations are correlated with patient prognoses and therapeutic resistance
- PMID: 30228934
- PMCID: PMC6140590
- DOI: 10.1080/2162402X.2018.1468951
Losses of cytokines and chemokines are common genetic features of human cancers: the somatic copy number alterations are correlated with patient prognoses and therapeutic resistance
Abstract
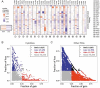
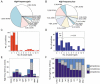
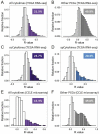
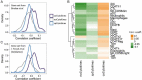
Intricate relationships among cytokines (including chemokines) shape the tumor microenvironment (TME) and reflect cell-cell interactions between malignant cells and other cells from the TME. Although our previous study indicated the transcriptional landscape of cytokines in 19 cancer types, the global pattern somatic copy number (SCN) alterations and the clinical relevance of cytokines have not been systematically investigated. Here, we reported a significant negative selection on cytokine genes. We also linked the SCN losses of cytokine genes to the abundance of immune infiltrates which affects cancer progression and patient prognoses. We also demonstrated and validated the correlations between SCN alterations of cytokine-containing loci and drug sensitivity. The results indicated the genomic loss of cytokines in malignant cells as a crucial theme for interrogating cancer progression, malignant cell-TME interactions, and therapeutics.
Keywords: cancer genetic landscape; chemokines; cytokines; pan-cancer bioinformatics analysis; somatic copy number (SCN) alterations.
Figures
Similar articles
-
Expression and clinical significance of CXC chemokines in the glioblastoma microenvironment.Life Sci. 2020 Nov 15;261:118486. doi: 10.1016/j.lfs.2020.118486. Epub 2020 Sep 23. Life Sci. 2020. PMID: 32976881
-
Characterization of cytokinome landscape for clinical responses in human cancers.Oncoimmunology. 2016 Nov 4;5(11):e1214789. doi: 10.1080/2162402X.2016.1214789. eCollection 2016. Oncoimmunology. 2016. PMID: 27999736 Free PMC article.
-
Specific immune cell and cytokine characteristics of human testicular germ cell neoplasia.Hum Reprod. 2016 Oct;31(10):2192-202. doi: 10.1093/humrep/dew211. Epub 2016 Sep 8. Hum Reprod. 2016. PMID: 27609978
-
Macrophage and Tumor Cell Cross-Talk Is Fundamental for Lung Tumor Progression: We Need to Talk.Front Oncol. 2020 Mar 11;10:324. doi: 10.3389/fonc.2020.00324. eCollection 2020. Front Oncol. 2020. PMID: 32219066 Free PMC article. Review.
-
[Analysis of genomic copy number alterations of malignant lymphomas and its application for diagnosis].Gan To Kagaku Ryoho. 2007 Jul;34(7):975-82. Gan To Kagaku Ryoho. 2007. PMID: 17637530 Review. Japanese.
Cited by
-
Melittin Induced G1 Cell Cycle Arrest and Apoptosis in Chago-K1 Human Bronchogenic Carcinoma Cells and Inhibited the Differentiation of THP-1 Cells into Tumour- Associated Macrophages.Asian Pac J Cancer Prev. 2018 Dec 25;19(12):3427-3434. doi: 10.31557/APJCP.2018.19.12.3427. Asian Pac J Cancer Prev. 2018. PMID: 30583665 Free PMC article.
-
Single-cell melanoma transcriptomes depicting functional versatility and clinical implications of STIM1 in the tumor microenvironment.Theranostics. 2021 Mar 5;11(11):5092-5106. doi: 10.7150/thno.54134. eCollection 2021. Theranostics. 2021. PMID: 33859736 Free PMC article.
-
GNAI2 Is a Risk Factor for Gastric Cancer: Study of Tumor Microenvironment (TME) and Establishment of Immune Risk Score (IRS).Oxid Med Cell Longev. 2022 Oct 14;2022:1254367. doi: 10.1155/2022/1254367. eCollection 2022. Oxid Med Cell Longev. 2022. Retraction in: Oxid Med Cell Longev. 2023 Sep 27;2023:9873973. doi: 10.1155/2023/9873973 PMID: 36275898 Free PMC article. Retracted.
-
The Role of Cytokinome in the HNSCC Tumor Microenvironment: A Narrative Review and Our Experience.Diagnostics (Basel). 2022 Nov 21;12(11):2880. doi: 10.3390/diagnostics12112880. Diagnostics (Basel). 2022. PMID: 36428939 Free PMC article. Review.
-
Antiangiogenesis Roles of Exosomes with Fei-Liu-Ping Ointment Treatment are Involved in the Lung Carcinoma with the Lewis Xenograft Mouse Model.Evid Based Complement Alternat Med. 2020 Apr 5;2020:9418593. doi: 10.1155/2020/9418593. eCollection 2020. Evid Based Complement Alternat Med. 2020. PMID: 32308722 Free PMC article.
References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
