Immediate-Early Promoter-Driven Transgenic Reporter System for Neuroethological Research in a Hemimetabolous Insect
- PMID: 30225346
- PMCID: PMC6140108
- DOI: 10.1523/ENEURO.0061-18.2018
Immediate-Early Promoter-Driven Transgenic Reporter System for Neuroethological Research in a Hemimetabolous Insect
Abstract
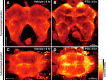
Genes expressed in response to increased neuronal activity are widely used as activity markers in recent behavioral neuroscience. In the present study, we established transgenic reporter system for whole-brain activity mapping in the two-spotted cricket Gryllus bimaculatus, a hemimetabolous insect used in neuroethology and behavioral ecology. In the cricket brain, a homolog of early growth response-1 (Gryllus egr-B) was rapidly induced as an immediate-early gene (IEG) in response to neuronal hyperexcitability. The upstream genomic fragment of Gryllus egr-B contains potential binding sites for transcription factors regulated by various intracellular signaling pathways, as well as core promoter elements conserved across insect/crustacean egr-B homologs. Using the upstream genomic fragment of Gryllus egr-B, we established an IEG promoter-driven transgenic reporter system in the cricket. In the brain of transgenic crickets, the reporter gene (a nuclear-targeted destabilized EYFP) was induced in response to neuronal hyperexcitability. Inducible expression of reporter protein was detected in almost all neurons after neuronal hyperexcitability. Using our novel reporter system, we successfully detected neuronal activation evoked by feeding in the cricket brain. Our IEG promoter-driven activity reporting system allows us to visualize behaviorally relevant neural circuits at cellular resolution in the cricket brain.
Keywords: Activity mapping; Gryllus bimaculatus; immediate-early gene; transgenesis.
Figures


Similar articles
-
GeneKnockout by Targeted Mutagenesis in a Hemimetabolous Insect, the Two-Spotted Cricket Gryllus bimaculatus, using TALENs.Methods Mol Biol. 2016;1338:143-55. doi: 10.1007/978-1-4939-2932-0_12. Methods Mol Biol. 2016. PMID: 26443220
-
Genome Editing in the Cricket, Gryllus bimaculatus.Methods Mol Biol. 2017;1630:219-233. doi: 10.1007/978-1-4939-7128-2_18. Methods Mol Biol. 2017. PMID: 28643262
-
Gene knockout by targeted mutagenesis in a hemimetabolous insect, the two-spotted cricket Gryllus bimaculatus, using TALENs.Methods. 2014 Aug 15;69(1):17-21. doi: 10.1016/j.ymeth.2014.05.006. Epub 2014 May 27. Methods. 2014. PMID: 24874787
-
The Cricket Gryllus bimaculatus: Techniques for Quantitative and Functional Genetic Analyses of Cricket Biology.Results Probl Cell Differ. 2019;68:183-216. doi: 10.1007/978-3-030-23459-1_8. Results Probl Cell Differ. 2019. PMID: 31598857 Review.
-
Activation and function of immediate-early genes in the nervous system.Biochem Cell Biol. 2011 Feb;89(1):61-73. doi: 10.1139/O10-138. Biochem Cell Biol. 2011. PMID: 21326363 Review.
Cited by
-
Design of Structural Parameters of Cutters for Tea Harvest Based on Biomimetic Methodology.Appl Bionics Biomech. 2021 Jul 22;2021:8798299. doi: 10.1155/2021/8798299. eCollection 2021. Appl Bionics Biomech. 2021. PMID: 34335873 Free PMC article.
-
Octopamine neurons mediate reward signals in social learning in an insect.iScience. 2023 Apr 8;26(5):106612. doi: 10.1016/j.isci.2023.106612. eCollection 2023 May 19. iScience. 2023. PMID: 37182108 Free PMC article.
-
Persistence of auditory modulation of wind-induced escape behavior in crickets.Front Physiol. 2023 May 9;14:1153913. doi: 10.3389/fphys.2023.1153913. eCollection 2023. Front Physiol. 2023. PMID: 37250114 Free PMC article.
-
Motor state changes escape behavior of crickets.iScience. 2023 Jul 11;26(8):107345. doi: 10.1016/j.isci.2023.107345. eCollection 2023 Aug 18. iScience. 2023. PMID: 37554465 Free PMC article.
-
Immediate early gene kakusei potentially plays a role in the daily foraging of honey bees.PLoS One. 2020 May 6;15(5):e0222256. doi: 10.1371/journal.pone.0222256. eCollection 2020. PLoS One. 2020. PMID: 32374761 Free PMC article.
References
-
- Adamo SA, Linn CE, Hoy RR (1995) The role of neurohormonal octopamine during ‘fight or flight’ behaviour in the field cricket Gryllus bimaculatus . J Exp Biol 198:1691–700. - PubMed
-
- Andersen CL, Jensen JL, Ørntoft TF (2004) Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res 64:5245–5250. 10.1158/0008-5472.CAN-04-0496 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
