The m6A-methylase complex recruits TREX and regulates mRNA export
- PMID: 30218090
- PMCID: PMC6138711
- DOI: 10.1038/s41598-018-32310-8
The m6A-methylase complex recruits TREX and regulates mRNA export
Abstract
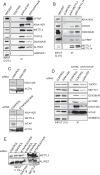
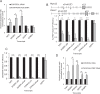
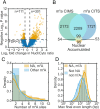
N6-methyladenosine (m6A) is the most abundant internal modification of eukaryotic mRNA. This modification has previously been shown to alter the export kinetics for mRNAs though the molecular details surrounding this phenomenon remain poorly understood. Recruitment of the TREX mRNA export complex to mRNA is driven by transcription, 5' capping and pre-mRNA splicing. Here we identify a fourth mechanism in human cells driving the association of TREX with mRNA involving the m6A methylase complex. We show that the m6A complex recruits TREX to m6A modified mRNAs and this process is essential for their efficient export. TREX also stimulates recruitment of the m6A reader protein YTHDC1 to the mRNA and the m6A complex influences the interaction of TREX with YTHDC1. Together our studies reveal a key role for TREX in the export of m6A modified mRNAs.
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
The m6A‑methylase complex and mRNA export.Biochim Biophys Acta Gene Regul Mech. 2019 Mar;1862(3):319-328. doi: 10.1016/j.bbagrm.2018.09.008. Epub 2018 Oct 2. Biochim Biophys Acta Gene Regul Mech. 2019. PMID: 30290229 Free PMC article. Review.
-
YTHDC1 mediates nuclear export of N6-methyladenosine methylated mRNAs.Elife. 2017 Oct 6;6:e31311. doi: 10.7554/eLife.31311. Elife. 2017. PMID: 28984244 Free PMC article.
-
TREX exposes the RNA-binding domain of Nxf1 to enable mRNA export.Nat Commun. 2012;3:1006. doi: 10.1038/ncomms2005. Nat Commun. 2012. PMID: 22893130 Free PMC article.
-
The human TREX-2 complex is stably associated with the nuclear pore basket.J Cell Sci. 2013 Jun 15;126(Pt 12):2656-67. doi: 10.1242/jcs.118000. Epub 2013 Apr 16. J Cell Sci. 2013. PMID: 23591820
-
TREX, SR proteins and export of mRNA.Curr Opin Cell Biol. 2005 Jun;17(3):269-73. doi: 10.1016/j.ceb.2005.04.011. Curr Opin Cell Biol. 2005. PMID: 15901496 Review.
Cited by
-
Overview of m6A and circRNAs in human cancers.J Cancer Res Clin Oncol. 2023 Aug;149(9):6769-6784. doi: 10.1007/s00432-023-04610-8. Epub 2023 Feb 18. J Cancer Res Clin Oncol. 2023. PMID: 36807759 Review.
-
The m6A‑methylase complex and mRNA export.Biochim Biophys Acta Gene Regul Mech. 2019 Mar;1862(3):319-328. doi: 10.1016/j.bbagrm.2018.09.008. Epub 2018 Oct 2. Biochim Biophys Acta Gene Regul Mech. 2019. PMID: 30290229 Free PMC article. Review.
-
MRT-ModSeq - Rapid detection of RNA modifications with MarathonRT.bioRxiv [Preprint]. 2023 May 25:2023.05.25.542276. doi: 10.1101/2023.05.25.542276. bioRxiv. 2023. Update in: J Mol Biol. 2023 Nov 15;435(22):168299. doi: 10.1016/j.jmb.2023.168299 PMID: 37292902 Free PMC article. Updated. Preprint.
-
Dynamic RNA methylation modifications and their regulatory role in mammalian development and diseases.Sci China Life Sci. 2024 Oct;67(10):2084-2104. doi: 10.1007/s11427-023-2526-2. Epub 2024 May 31. Sci China Life Sci. 2024. PMID: 38833084 Review.
-
mRNA Regulation by RNA Modifications.Annu Rev Biochem. 2023 Jun 20;92:175-198. doi: 10.1146/annurev-biochem-052521-035949. Epub 2023 Apr 5. Annu Rev Biochem. 2023. PMID: 37018844 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

