Cytosolic Recognition of Microbes and Pathogens: Inflammasomes in Action
- PMID: 30209070
- PMCID: PMC6298609
- DOI: 10.1128/MMBR.00015-18
Cytosolic Recognition of Microbes and Pathogens: Inflammasomes in Action
Abstract
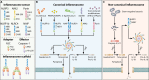
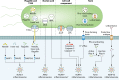
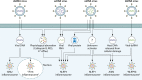
Infection is a dynamic biological process underpinned by a complex interplay between the pathogen and the host. Microbes from all domains of life, including bacteria, viruses, fungi, and protozoan parasites, have the capacity to cause infection. Infection is sensed by the host, which often leads to activation of the inflammasome, a cytosolic macromolecular signaling platform that mediates the release of the proinflammatory cytokines interleukin-1β (IL-1β) and IL-18 and cleavage of the pore-forming protein gasdermin D, leading to pyroptosis. Host-mediated sensing of the infection occurs when pathogens inject or carry pathogen-associated molecular patterns (PAMPs) into the cytoplasm or induce damage that causes cytosolic liberation of danger-associated molecular patterns (DAMPs) in the host cell. Recognition of PAMPs and DAMPs by inflammasome sensors, including NLRP1, NLRP3, NLRC4, NAIP, AIM2, and Pyrin, initiates a cascade of events that culminate in inflammation and cell death. However, pathogens can deploy virulence factors capable of minimizing or evading host detection. This review presents a comprehensive overview of the mechanisms of microbe-induced activation of the inflammasome and the functional consequences of inflammasome activation in infectious diseases. We also explore the microbial strategies used in the evasion of inflammasome sensing at the host-microbe interaction interface.
Keywords: AIM2; NAIP; NLRC4; NLRP1; NLRP3; Pyrin; caspase-1; caspase-11; gasdermin; pyroptosis.
Copyright © 2018 American Society for Microbiology.
Figures
Similar articles
-
Advances in Understanding Activation and Function of the NLRC4 Inflammasome.Int J Mol Sci. 2021 Jan 21;22(3):1048. doi: 10.3390/ijms22031048. Int J Mol Sci. 2021. PMID: 33494299 Free PMC article. Review.
-
Cytokine Secretion and Pyroptosis of Thyroid Follicular Cells Mediated by Enhanced NLRP3, NLRP1, NLRC4, and AIM2 Inflammasomes Are Associated With Autoimmune Thyroiditis.Front Immunol. 2018 Jun 4;9:1197. doi: 10.3389/fimmu.2018.01197. eCollection 2018. Front Immunol. 2018. PMID: 29915579 Free PMC article.
-
IRGB10 Liberates Bacterial Ligands for Sensing by the AIM2 and Caspase-11-NLRP3 Inflammasomes.Cell. 2016 Oct 6;167(2):382-396.e17. doi: 10.1016/j.cell.2016.09.012. Epub 2016 Sep 29. Cell. 2016. PMID: 27693356 Free PMC article.
-
Inflammasomes and its importance in viral infections.Immunol Res. 2016 Dec;64(5-6):1101-1117. doi: 10.1007/s12026-016-8873-z. Immunol Res. 2016. PMID: 27699580 Review.
-
Emerging Activators and Regulators of Inflammasomes and Pyroptosis.Trends Immunol. 2019 Nov;40(11):1035-1052. doi: 10.1016/j.it.2019.09.005. Epub 2019 Oct 29. Trends Immunol. 2019. PMID: 31662274 Review.
Cited by
-
AIM2 Inflammasome's First Decade of Discovery: Focus on Oral Diseases.Front Immunol. 2020 Aug 13;11:1487. doi: 10.3389/fimmu.2020.01487. eCollection 2020. Front Immunol. 2020. PMID: 32903550 Free PMC article. Review.
-
NLRP1 in Cutaneous SCCs: An Example of the Complex Roles of Inflammasomes in Cancer Development.Int J Mol Sci. 2022 Oct 14;23(20):12308. doi: 10.3390/ijms232012308. Int J Mol Sci. 2022. PMID: 36293159 Free PMC article. Review.
-
Irgm2 and Gate-16 cooperatively dampen Gram-negative bacteria-induced caspase-11 response.EMBO Rep. 2020 Nov 5;21(11):e50829. doi: 10.15252/embr.202050829. Epub 2020 Oct 30. EMBO Rep. 2020. PMID: 33124769 Free PMC article.
-
Inflammasome activation and regulation: toward a better understanding of complex mechanisms.Cell Discov. 2020 Jun 9;6:36. doi: 10.1038/s41421-020-0167-x. eCollection 2020. Cell Discov. 2020. PMID: 32550001 Free PMC article. Review.
-
Interaction of Mycobacteria With Host Cell Inflammasomes.Front Immunol. 2022 Feb 14;13:791136. doi: 10.3389/fimmu.2022.791136. eCollection 2022. Front Immunol. 2022. PMID: 35237260 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

