Dynamic Interplay of RNA and Protein in the Human Immunodeficiency Virus-1 Reverse Transcription Initiation Complex
- PMID: 30201267
- PMCID: PMC6289658
- DOI: 10.1016/j.jmb.2018.08.029
Dynamic Interplay of RNA and Protein in the Human Immunodeficiency Virus-1 Reverse Transcription Initiation Complex
Abstract
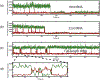
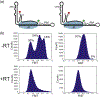
The initiation of reverse transcription in human immunodeficiency virus-1 is a key early step in the virus replication cycle. During this process, the viral enzyme reverse transcriptase (RT) copies the single-stranded viral RNA (vRNA) genome into double-stranded DNA using human tRNALys3 as a primer for initiation. The tRNA primer and vRNA genome contain several complementary sequences that are important for regulating reverse transcription initiation kinetics. Using single-molecule Förster resonance energy transfer spectroscopy, we demonstrate that the vRNA-tRNA initiation complex is conformationally heterogeneous and dynamic in the absence of RT. As shown previously, nucleic acid-RT interaction is characterized by rapid dissociation constants. We show that extension of the vRNA-tRNA primer binding site helix from 18 base pairs to 22 base pairs stabilizes RT binding to the complex and that the tRNA 5' end has a role in modulating RT binding. RT occupancy on the complex stabilizes helix 1 formation and reduces global structural heterogeneity. The stabilization of helix 1 upon RT binding may serve to destabilize helix 2, the first pause site for RT during initiation, during later steps of reverse transcription initiation.
Keywords: HIV-1; RNA dynamics; protein and RNA interactions; reverse transcriptase; single-molecule FRET.
Copyright © 2018 Elsevier Ltd. All rights reserved.
Figures


Similar articles
-
MD simulations of HIV-1 RT primer-template complex: effect of modified nucleosides and antisense PNA oligomer.J Biomol Struct Dyn. 2013;31(6):539-60. doi: 10.1080/07391102.2012.706076. Epub 2012 Aug 13. J Biomol Struct Dyn. 2013. PMID: 22888964
-
Forced selection of a human immunodeficiency virus type 1 variant that uses a non-self tRNA primer for reverse transcription: involvement of viral RNA sequences and the reverse transcriptase enzyme.J Virol. 2004 Oct;78(19):10706-14. doi: 10.1128/JVI.78.19.10706-10714.2004. J Virol. 2004. PMID: 15367637 Free PMC article.
-
Direct and indirect contributions of RNA secondary structure elements to the initiation of HIV-1 reverse transcription.J Biol Chem. 2002 Nov 8;277(45):43233-42. doi: 10.1074/jbc.M205295200. Epub 2002 Aug 22. J Biol Chem. 2002. PMID: 12194974
-
HIV-1 reverse transcription initiation: a potential target for novel antivirals?Virus Res. 2008 Jun;134(1-2):4-18. doi: 10.1016/j.virusres.2007.12.009. Epub 2008 Feb 6. Virus Res. 2008. PMID: 18255184 Review.
-
Initiation of HIV-1 reverse transcription and functional role of nucleocapsid-mediated tRNA/viral genome interactions.Virus Res. 2012 Nov;169(2):324-39. doi: 10.1016/j.virusres.2012.06.006. Epub 2012 Jun 18. Virus Res. 2012. PMID: 22721779 Review.
Cited by
-
Expanding single-molecule fluorescence spectroscopy to capture complexity in biology.Curr Opin Struct Biol. 2019 Oct;58:233-240. doi: 10.1016/j.sbi.2019.05.005. Epub 2019 Jun 15. Curr Opin Struct Biol. 2019. PMID: 31213390 Free PMC article. Review.
-
Advances in understanding the initiation of HIV-1 reverse transcription.Curr Opin Struct Biol. 2020 Dec;65:175-183. doi: 10.1016/j.sbi.2020.07.005. Epub 2020 Sep 8. Curr Opin Struct Biol. 2020. PMID: 32916568 Free PMC article. Review.
-
Illuminating the virus life cycle with single-molecule FRET imaging.Adv Virus Res. 2019;105:239-273. doi: 10.1016/bs.aivir.2019.07.004. Epub 2019 Aug 20. Adv Virus Res. 2019. PMID: 31522706 Free PMC article.
-
Distinct Conformational States Underlie Pausing during Initiation of HIV-1 Reverse Transcription.J Mol Biol. 2020 Jul 24;432(16):4499-4522. doi: 10.1016/j.jmb.2020.06.003. Epub 2020 Jun 6. J Mol Biol. 2020. PMID: 32512005 Free PMC article.
-
Structure of HIV-1 RT/dsRNA initiation complex prior to nucleotide incorporation.Proc Natl Acad Sci U S A. 2019 Apr 9;116(15):7308-7313. doi: 10.1073/pnas.1814170116. Epub 2019 Mar 22. Proc Natl Acad Sci U S A. 2019. PMID: 30902895 Free PMC article.
References
-
- Baltimore D (1970). RNA-dependent DNA polymerase in virions of RNA tumour viruses. Nature 226, 1209–11. - PubMed
-
- Temin HM & Mizutani S (1970). RNA-dependent DNA polymerase in virions of Rous sarcoma virus. Nature 226, 1211–3. - PubMed
-
- Haseltine WA, Kleid DG, Panet A, Rothenberg E & Baltimore D (1976). Ordered Transcription of Rna Tumor-Virus Genomes. Journal of Molecular Biology 106, 109–131. - PubMed
-
- Mitsuya H, Weinhold KJ, Furman PA, Stclair MH, Lehrman SN, Gallo RC, Bolognesi D, Barry DW & Broder S (1985). 3’-Azido-3’-Deoxythymidine (Bw A509u) - an Antiviral Agent That Inhibits the Infectivity and Cytopathic Effect of Human Lymphotropic-T Virus Type-Iii Lymphadenopathy-Associated Virus Invitro. Proceedings of the National Academy of Sciences of the United States of America 82, 7096–7100. - PMC - PubMed
-
- Furman PA, Fyfe JA, Stclair MH, Weinhold K, Rideout JL, Freeman GA, Lehrman SN, Bolognesi DP, Broder S, Mitsuya H & Barry DW (1986). Phosphorylation of 3’-Azido-3’- Deoxythymidine and Selective Interaction of the 5’-Triphosphate with HumanImmunodeficiencyVirus Reverse-Transcriptase. Proceedings of the National Academy of Sciences of the United States of America 83, 8333–8337. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

