DNA methylation footprints during soybean domestication and improvement
- PMID: 30201012
- PMCID: PMC6130073
- DOI: 10.1186/s13059-018-1516-z
DNA methylation footprints during soybean domestication and improvement
Abstract
Background: In addition to genetic variation, epigenetic variation plays an important role in determining various biological processes. The importance of natural genetic variation to crop domestication and improvement has been widely investigated. However, the contribution of epigenetic variation in crop domestication at population level has rarely been explored.
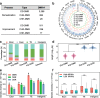
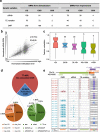
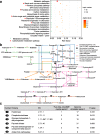
Results: To understand the impact of epigenetics on crop domestication, we investigate the variation of DNA methylation during soybean domestication and improvement by whole-genome bisulfite sequencing of 45 soybean accessions, including wild soybeans, landraces, and cultivars. Through methylomic analysis, we identify 5412 differentially methylated regions (DMRs). These DMRs exhibit characters distinct from those of genetically selected regions. In particular, they have significantly higher genetic diversity. Association analyses suggest only 22.54% of DMRs can be explained by local genetic variations. Intriguingly, genes in the DMRs that are not associated with any genetic variation are enriched in carbohydrate metabolism pathways.
Conclusions: This study provides a valuable map of DNA methylation across diverse accessions and dissects the relationship between DNA methylation variation and genetic variation during soybean domestication, thus expanding our understanding of soybean domestication and improvement.
Keywords: DNA methylation; Domestication; Soybean.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures


Similar articles
-
Population analysis reveals the roles of DNA methylation in tomato domestication and metabolic diversity.Sci China Life Sci. 2023 Aug;66(8):1888-1902. doi: 10.1007/s11427-022-2299-5. Epub 2023 Mar 23. Sci China Life Sci. 2023. PMID: 36971992
-
Increased DNA methylation contributes to the early ripening of pear fruits during domestication and improvement.Genome Biol. 2024 Apr 5;25(1):87. doi: 10.1186/s13059-024-03220-y. Genome Biol. 2024. PMID: 38581061 Free PMC article.
-
Genetic diversity patterns and domestication origin of soybean.Theor Appl Genet. 2019 Apr;132(4):1179-1193. doi: 10.1007/s00122-018-3271-7. Epub 2018 Dec 26. Theor Appl Genet. 2019. PMID: 30588539 Free PMC article.
-
Soybean domestication: the origin, genetic architecture and molecular bases.New Phytol. 2017 Apr;214(2):539-553. doi: 10.1111/nph.14418. Epub 2017 Jan 30. New Phytol. 2017. PMID: 28134435 Review.
-
Recent developments of genomic research in soybean.J Genet Genomics. 2012 Jul 20;39(7):317-24. doi: 10.1016/j.jgg.2012.02.002. Epub 2012 Feb 18. J Genet Genomics. 2012. PMID: 22835978 Review.
Cited by
-
Altered chromatin architecture and gene expression during polyploidization and domestication of soybean.Plant Cell. 2021 Jul 2;33(5):1430-1446. doi: 10.1093/plcell/koab081. Plant Cell. 2021. PMID: 33730165 Free PMC article.
-
Epigenetic gene regulation in plants and its potential applications in crop improvement.Nat Rev Mol Cell Biol. 2025 Jan;26(1):51-67. doi: 10.1038/s41580-024-00769-1. Epub 2024 Aug 27. Nat Rev Mol Cell Biol. 2025. PMID: 39192154 Review.
-
DNA methylation analysis reveals local changes in resistant and susceptible soybean lines in response to Phytophthora sansomeana.G3 (Bethesda). 2024 Oct 7;14(10):jkae191. doi: 10.1093/g3journal/jkae191. G3 (Bethesda). 2024. PMID: 39141590 Free PMC article.
-
Regulation of seed traits in soybean.aBIOTECH. 2023 Nov 27;4(4):372-385. doi: 10.1007/s42994-023-00122-8. eCollection 2023 Dec. aBIOTECH. 2023. PMID: 38106437 Free PMC article. Review.
-
Genome-Wide Identification, Expression and Interaction Analysis of GmSnRK2 and Type A PP2C Genes in Response to Abscisic Acid Treatment and Drought Stress in Soybean Plant.Int J Mol Sci. 2022 Oct 29;23(21):13166. doi: 10.3390/ijms232113166. Int J Mol Sci. 2022. PMID: 36361951 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

