The Protein Coded by a Short Open Reading Frame, Not by the Annotated Coding Sequence, Is the Main Gene Product of the Dual-Coding Gene MIEF1
- PMID: 30181344
- PMCID: PMC6283296
- DOI: 10.1074/mcp.RA118.000593
The Protein Coded by a Short Open Reading Frame, Not by the Annotated Coding Sequence, Is the Main Gene Product of the Dual-Coding Gene MIEF1
Abstract
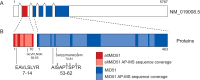
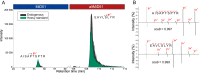
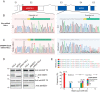
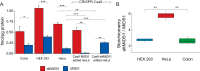
Proteogenomics and ribosome profiling concurrently show that genes may code for both a large and one or more small proteins translated from annotated coding sequences (CDSs) and unannotated alternative open reading frames (named alternative ORFs or altORFs), respectively, but the stoichiometry between large and small proteins translated from a same gene is unknown. MIEF1, a gene recently identified as a dual-coding gene, harbors a CDS and a newly annotated and actively translated altORF located in the 5'UTR. Here, we use absolute quantification with stable isotope-labeled peptides and parallel reaction monitoring to determine levels of both proteins in two human cells lines and in human colon. We report that the main MIEF1 translational product is not the canonical 463 amino acid MiD51 protein but the small 70 amino acid alternative MiD51 protein (altMiD51). These results demonstrate the inadequacy of the single CDS concept and provide a strong argument for incorporating altORFs and small proteins in functional annotations.
Keywords: Absolute quantification; Gene Expression*; Knockouts*; Mass Spectrometry; Mitochondria function or biology; Parallel reaction monitoring; Proteogenomics; Translation*; alternative translation; short ORF.
© 2018 Delcourt et al.
Figures
Similar articles
-
Deep transcriptome annotation enables the discovery and functional characterization of cryptic small proteins.Elife. 2017 Oct 30;6:e27860. doi: 10.7554/eLife.27860. Elife. 2017. PMID: 29083303 Free PMC article.
-
MIEF1 Microprotein Regulates Mitochondrial Translation.Biochemistry. 2018 Sep 25;57(38):5564-5575. doi: 10.1021/acs.biochem.8b00726. Epub 2018 Sep 14. Biochemistry. 2018. PMID: 30215512 Free PMC article.
-
Direct detection of alternative open reading frames translation products in human significantly expands the proteome.PLoS One. 2013 Aug 12;8(8):e70698. doi: 10.1371/journal.pone.0070698. eCollection 2013. PLoS One. 2013. PMID: 23950983 Free PMC article.
-
Small Proteins Encoded by Unannotated ORFs are Rising Stars of the Proteome, Confirming Shortcomings in Genome Annotations and Current Vision of an mRNA.Proteomics. 2018 May;18(10):e1700058. doi: 10.1002/pmic.201700058. Epub 2017 Oct 11. Proteomics. 2018. PMID: 28627015 Review.
-
Translation of Small Open Reading Frames: Roles in Regulation and Evolutionary Innovation.Trends Genet. 2019 Mar;35(3):186-198. doi: 10.1016/j.tig.2018.12.003. Epub 2018 Dec 31. Trends Genet. 2019. PMID: 30606460 Review.
Cited by
-
Noncanonical altPIDD1 protein: unveiling the true major translational output of the PIDD1 gene.Life Sci Alliance. 2024 Nov 12;8(2):e202402910. doi: 10.26508/lsa.202402910. Print 2025 Feb. Life Sci Alliance. 2024. PMID: 39532532 Free PMC article.
-
High Polymorphism Levels of De Novo ORFs in a Yoruba Human Population.Genome Biol Evol. 2024 Jul 3;16(7):evae126. doi: 10.1093/gbe/evae126. Genome Biol Evol. 2024. PMID: 38934859 Free PMC article.
-
Space radiation damage rescued by inhibition of key spaceflight associated miRNAs.Nat Commun. 2024 Jun 11;15(1):4825. doi: 10.1038/s41467-024-48920-y. Nat Commun. 2024. PMID: 38862542 Free PMC article.
-
An Inner Mitochondrial Membrane Microprotein from the SLC35A4 Upstream ORF Regulates Cellular Metabolism.J Mol Biol. 2024 May 15;436(10):168559. doi: 10.1016/j.jmb.2024.168559. Epub 2024 Apr 3. J Mol Biol. 2024. PMID: 38580077 Free PMC article.
-
Heimdall, an alternative protein issued from a ncRNA related to kappa light chain variable region of immunoglobulins from astrocytes: a new player in neural proteome.Cell Death Dis. 2023 Aug 16;14(8):526. doi: 10.1038/s41419-023-06037-y. Cell Death Dis. 2023. PMID: 37587118 Free PMC article.
References
-
- Vanderperre B., Lucier J. F., Bissonnette C., Motard J., Tremblay G., Vanderperre S., Wisztorski M., Salzet M., Boisvert F.M., and Roucou X. (2013) Direct detection of alternative open reading frames translation products in human significantly expands the proteome. PloS ONE, 8, e70698. - PMC - PubMed
-
- Menschaert G., Van Criekinge W., Notelaers T., Koch A., Crappé J., Gevaert K., and Van Damme P. (2013) Deep proteome coverage based on ribosome profiling aids mass spectrometry-based protein and peptide discovery and provides evidence of alternative translation products and near-cognate translation initiation events. Mol. Cell. Proteomics 12, 1780–1790 - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

