Transcriptomic Insights Into the Growth Phase- and Sugar-Associated Changes in the Exopolysaccharide Production of a High EPS-Producing Streptococcus thermophilus ASCC 1275
- PMID: 30177921
- PMCID: PMC6109772
- DOI: 10.3389/fmicb.2018.01919
Transcriptomic Insights Into the Growth Phase- and Sugar-Associated Changes in the Exopolysaccharide Production of a High EPS-Producing Streptococcus thermophilus ASCC 1275
Abstract
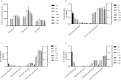
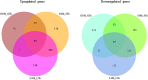
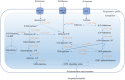
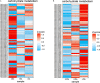
In a previous study, incorporation of high exopolysaccharide (EPS) producing dairy starter bacterium Streptococcus thermophilus ASCC 1275 was found to improve functionality of low fat mozzarella cheese and yogurt. This bacterium in its eps gene cluster has a unique pair of chain length determining genes, epsC- epsD, when compared to other sequenced S. thermophilus strains. Hence, the aim of this study was to understand the regulatory mechanism of EPS production in this bacterium using transcriptomic analysis to provide opportunities to improve the yield of EPS. As sugars are considered as one of the major determinants of EPS production, after preliminary screening, we selected three sugars, glucose, sucrose and lactose to identify the EPS producing mechanism of this bacterium in M17 medium. Complete RNA-seq analysis was performed using Illumina HiSeq 2000 sequencing system on S. thermophilus 1275 grown in three different sugars at two-time points, 5 h (log phase) and 10 h (stationary phase) to recognize the genes involved in sugar uptake, UDP-sugar formation, EPS assembly and export of EPS outside the bacterial cell. S. thermophilus 1275 was found to produce high amount of EPS (∼430 mg/L) in sucrose (1%) supplemented M17 medium when compared to other two sugars. Differential gene expression analysis revealed the involvement of phosphoenolpyruvate phosphotransferase system (PEP-PTS) for glucose and sucrose uptake, and lacS gene for lactose uptake. The pathways for the formation of UDP-glucose and UDP-galactose were highly upregulated in all the three sugars. In the presence of sucrose, eps1C1D2C2D were found to be highly expressed which refers to high EPS production. Protein homology study suggested the presence of Wzx/Wzy-dependent EPS synthesis and transport pathway in this bacterium. KEGG pathway and COG functional enrichment analysis were also performed to support the result. This is the first report providing the transcriptomic insights into the EPS production mechanism of a common dairy bacterium, S. thermophilus.
Keywords: RNA-seq; S. thermophilus ASCC 1275; exopolysaccharide; sugars; transcriptomics.
Figures




Similar articles
-
Proteomic analysis reveals potential factors associated with enhanced EPS production in Streptococcus thermophilus ASCC 1275.Sci Rep. 2020 Jan 21;10(1):807. doi: 10.1038/s41598-020-57665-9. Sci Rep. 2020. PMID: 31964939 Free PMC article.
-
Genomic insights into high exopolysaccharide-producing dairy starter bacterium Streptococcus thermophilus ASCC 1275.Sci Rep. 2014 May 15;4:4974. doi: 10.1038/srep04974. Sci Rep. 2014. PMID: 24827399 Free PMC article.
-
Genomic and phenotypic analyses of exopolysaccharide biosynthesis in Streptococcus thermophilus S-3.J Dairy Sci. 2019 Jun;102(6):4925-4934. doi: 10.3168/jds.2018-15572. Epub 2019 Mar 28. J Dairy Sci. 2019. PMID: 30928267
-
Biochemistry, genetics, and applications of exopolysaccharide production in Streptococcus thermophilus: a review.J Dairy Sci. 2003 Feb;86(2):407-23. doi: 10.3168/jds.S0022-0302(03)73619-4. J Dairy Sci. 2003. PMID: 12647947 Review.
-
New advances in exopolysaccharides production of Streptococcus thermophilus.Arch Microbiol. 2017 Aug;199(6):799-809. doi: 10.1007/s00203-017-1366-1. Epub 2017 Mar 29. Arch Microbiol. 2017. PMID: 28357474 Review.
Cited by
-
Transcriptome Analysis Reveals the Role of Sucrose in the Production of Latilactobacillus sakei L3 Exopolysaccharide.Int J Mol Sci. 2024 Jun 29;25(13):7185. doi: 10.3390/ijms25137185. Int J Mol Sci. 2024. PMID: 39000292 Free PMC article.
-
Proteomic analysis reveals potential factors associated with enhanced EPS production in Streptococcus thermophilus ASCC 1275.Sci Rep. 2020 Jan 21;10(1):807. doi: 10.1038/s41598-020-57665-9. Sci Rep. 2020. PMID: 31964939 Free PMC article.
-
Omics Approaches to Assess Flavor Development in Cheese.Foods. 2022 Jan 11;11(2):188. doi: 10.3390/foods11020188. Foods. 2022. PMID: 35053920 Free PMC article. Review.
-
Lactic Acid Bacteria Exopolysaccharides Producers: A Sustainable Tool for Functional Foods.Foods. 2021 Jul 17;10(7):1653. doi: 10.3390/foods10071653. Foods. 2021. PMID: 34359523 Free PMC article. Review.
-
Genome-Scale Metabolic Modeling Combined with Transcriptome Profiling Provides Mechanistic Understanding of Streptococcus thermophilus CH8 Metabolism.Appl Environ Microbiol. 2022 Aug 23;88(16):e0078022. doi: 10.1128/aem.00780-22. Epub 2022 Aug 4. Appl Environ Microbiol. 2022. PMID: 35924931 Free PMC article.
References
-
- Bhaskaracharya R., Shah N. P. (2000). Texture characteristics and microstructure of skim milk mozzarella cheeses made using exopolysaccharide or non-exopolysaccharide producing starter cultures. Aust. J. Dairy Technol. 55:132.
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

