STAT3 Cooperates With Phospholipid Scramblase 2 to Suppress Type I Interferon Response
- PMID: 30158934
- PMCID: PMC6104169
- DOI: 10.3389/fimmu.2018.01886
STAT3 Cooperates With Phospholipid Scramblase 2 to Suppress Type I Interferon Response
Abstract
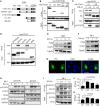
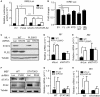
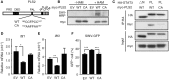
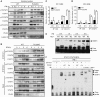
Type I interferon (IFN-I) is a pluripotent cytokine that modulates innate and adaptive immunity. We have previously shown that STAT3 suppresses IFN-I response in a manner dependent on its N-terminal domain (NTD), but independent of its DNA-binding and transactivation ability. Using the yeast two-hybrid system, we have identified phospholipid scramblase 2 (PLSCR2) as a STAT3 NTD-binding partner and a suppressor of IFN-I response. Overexpression of PLSCR2 attenuates ISRE-driven reporter activity, which is further aggravated by co-expression of STAT3. Moreover, PLSCR2 deficiency enhances IFN-I-induced gene expression and antiviral activity without affecting the activation or nuclear translocation of STAT1 and STAT2 or the assembly of ISGF3 complex. Instead, PLSCR2 impedes promoter occupancy by ISGF3, an effect further intensified by the presence of STAT3. Moreover, palmitoylation of PLSCR2 is required for its binding to STAT3 and for this suppressive activity. In addition to STAT3, PLSCR2 also interacts with STAT2, which facilitates the suppressive effect on ISGF3-mediated transcriptional activity. Together, these results define the role of a novel STAT3-PLSCR2 axis in fine-tuning IFN-I response.
Keywords: IFN-stimulated gene; STAT3; palmitoylation; phospholipid scramblase 2; type I interferon.
Figures


Similar articles
-
Fine-Tuning of Type I Interferon Response by STAT3.Front Immunol. 2019 Jun 26;10:1448. doi: 10.3389/fimmu.2019.01448. eCollection 2019. Front Immunol. 2019. PMID: 31293595 Free PMC article. Review.
-
Opposed regulation of type I IFN-induced STAT3 and ISGF3 transcriptional activities by histone deacetylases (HDACS) 1 and 2.FASEB J. 2012 Jan;26(1):240-9. doi: 10.1096/fj.11-191122. Epub 2011 Sep 28. FASEB J. 2012. PMID: 21957129
-
The unique role of STAT2 in constitutive and IFN-induced transcription and antiviral responses.Cytokine Growth Factor Rev. 2016 Jun;29:71-81. doi: 10.1016/j.cytogfr.2016.02.010. Epub 2016 Mar 18. Cytokine Growth Factor Rev. 2016. PMID: 27053489 Review.
-
Interferon-gamma inhibits interferon-alpha signalling in hepatic cells: evidence for the involvement of STAT1 induction and hyperexpression of STAT1 in chronic hepatitis C.Biochem J. 2004 Apr 1;379(Pt 1):199-208. doi: 10.1042/BJ20031495. Biochem J. 2004. PMID: 14690454 Free PMC article.
-
An RNA biding protein, Y14 interacts with and modulates STAT3 activation.Biochem Biophys Res Commun. 2008 Aug 1;372(3):475-9. doi: 10.1016/j.bbrc.2008.05.073. Epub 2008 May 27. Biochem Biophys Res Commun. 2008. PMID: 18503751
Cited by
-
Lyssavirus P-protein selectively targets STAT3-STAT1 heterodimers to modulate cytokine signalling.PLoS Pathog. 2020 Sep 9;16(9):e1008767. doi: 10.1371/journal.ppat.1008767. eCollection 2020 Sep. PLoS Pathog. 2020. PMID: 32903273 Free PMC article.
-
The Molecular Basis of Viral Inhibition of IRF- and STAT-Dependent Immune Responses.Front Immunol. 2019 Jan 8;9:3086. doi: 10.3389/fimmu.2018.03086. eCollection 2018. Front Immunol. 2019. PMID: 30671058 Free PMC article. Review.
-
Phospholipid Scramblases: Role in Cancer Progression and Anticancer Therapeutics.Front Genet. 2022 Mar 29;13:875894. doi: 10.3389/fgene.2022.875894. eCollection 2022. Front Genet. 2022. PMID: 35422844 Free PMC article. Review.
-
Capsaicin functions as a selective degrader of STAT3 to enhance host resistance to viral infection.Acta Pharmacol Sin. 2023 Nov;44(11):2253-2264. doi: 10.1038/s41401-023-01111-9. Epub 2023 Jun 13. Acta Pharmacol Sin. 2023. PMID: 37311796 Free PMC article.
-
The Dynamic Interface of Viruses with STATs.J Virol. 2020 Oct 27;94(22):e00856-20. doi: 10.1128/JVI.00856-20. Print 2020 Oct 27. J Virol. 2020. PMID: 32847860 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

