A novel molecular rotor facilitates detection of p53-DNA interactions using the Fluorescent Intercalator Displacement Assay
- PMID: 30154420
- PMCID: PMC6113202
- DOI: 10.1038/s41598-018-31197-9
A novel molecular rotor facilitates detection of p53-DNA interactions using the Fluorescent Intercalator Displacement Assay
Abstract
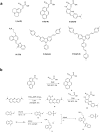
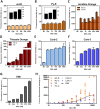
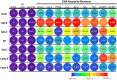
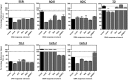
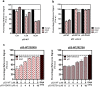
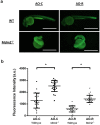
We have investigated the use of fluorescent molecular rotors as probes for detection of p53 binding to DNA. These are a class of fluorophores that undergo twisted intramolecular charge transfer (TICT). They are non-fluorescent in a freely rotating conformation and experience a fluorescence increase when restricted in the planar conformation. We hypothesized that intercalation of a molecular rotor between DNA base pairs would result in a fluorescence turn-on signal. Upon displacement by a DNA binding protein, measurable loss of signal would facilitate use of the molecular rotor in the fluorescent intercalator displacement (FID) assay. A panel of probes was interrogated using the well-established p53 model system across various DNA response elements. A novel, readily synthesizable molecular rotor incorporating an acridine orange DNA intercalating group (AO-R) outperformed other conventional dyes in the FID assay. It enabled relative measurement of p53 sequence-specific DNA interactions and study of the dominant-negative effects of cancer-associated p53 mutants. In a further application, AO-R also proved useful for staining apoptotic cells in live zebrafish embryos.
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
A fluorescent intercalator displacement assay for establishing DNA binding selectivity and affinity.Curr Protoc Nucleic Acid Chem. 2005 Apr;Chapter 8:Unit 8.5. doi: 10.1002/0471142700.nc0805s20. Curr Protoc Nucleic Acid Chem. 2005. PMID: 18428943
-
Identification of new DNA i-motif binding ligands through a fluorescent intercalator displacement assay.Org Biomol Chem. 2017 Jul 21;15(27):5669-5673. doi: 10.1039/c7ob00710h. Epub 2017 Jun 1. Org Biomol Chem. 2017. PMID: 28567459 Free PMC article.
-
Thiazole orange as the fluorescent intercalator in a high resolution fid assay for determining DNA binding affinity and sequence selectivity of small molecules.Bioorg Med Chem. 2001 Sep;9(9):2511-8. doi: 10.1016/s0968-0896(01)00243-7. Bioorg Med Chem. 2001. PMID: 11553493
-
Harnessing DNA intercalation.Trends Biotechnol. 2007 Oct;25(10):433-6. doi: 10.1016/j.tibtech.2007.08.003. Epub 2007 Sep 7. Trends Biotechnol. 2007. PMID: 17825446 Review.
-
Excited-state photophysics of an acridine derivative selectively intercalated in duplex DNA.Chemphyschem. 2002 May 17;3(5):452-5. doi: 10.1002/1439-7641(20020517)3:5<452::AID-CPHC452>3.0.CO;2-K. Chemphyschem. 2002. PMID: 12465507 Review. No abstract available.
Cited by
-
Tracking Small Extracellular Vesicles Using a Minimally Invasive PicoGreen Labeling Strategy.ACS Appl Bio Mater. 2024 Nov 18;7(11):7770-7783. doi: 10.1021/acsabm.4c01500. Epub 2024 Oct 31. ACS Appl Bio Mater. 2024. PMID: 39482871 Free PMC article.
References
MeSH terms
Substances
Grants and funding
- 11/03/EG/07/01/Agency for Science, Technology and Research (A*STAR)/International
- 11/03/EG/07/01/Agency for Science, Technology and Research (A*STAR)/International
- 11/03/EG/07/01/Agency for Science, Technology and Research (A*STAR)/International
- 11/03/EG/07/01/Agency for Science, Technology and Research (A*STAR)/International
- 11/03/EG/07/01/Agency for Science, Technology and Research (A*STAR)/International
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

