Nitric Oxide Modulates Endonuclease III Redox Activity by a 800 mV Negative Shift upon [Fe4S4] Cluster Nitrosylation
- PMID: 30145881
- PMCID: PMC6186442
- DOI: 10.1021/jacs.8b07362
Nitric Oxide Modulates Endonuclease III Redox Activity by a 800 mV Negative Shift upon [Fe4S4] Cluster Nitrosylation
Abstract
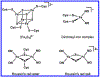
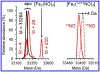
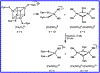
Here we characterize the [Fe4S4] cluster nitrosylation of a DNA repair enzyme, endonuclease III (EndoIII), using DNA-modified gold electrochemistry and protein film voltammetry, electrophoretic mobility shift assays, mass spectrometry of whole and trypsin-digested protein, and a variety of spectroscopies. Exposure of EndoIII to nitric oxide under anaerobic conditions transforms the [Fe4S4] cluster into a dinitrosyl iron complex, [(Cys)2Fe(NO)2]-, and Roussin's red ester, [(μ-Cys)2Fe2(NO)4], in a 1:1 ratio with an average retention of 3.05 ± 0.01 Fe per nitrosylated cluster. The formation of the dinitrosyl iron complex is consistent with previous reports, but the Roussin's red ester is an unreported product of EndoIII nitrosylation. Hyperfine sublevel correlation (HYSCORE) pulse EPR spectroscopy detects two distinct classes of NO with 14N hyperfine couplings consistent with the dinitrosyl iron complex and reduced Roussin's red ester. Whole-protein mass spectrometry of EndoIII nitrosylated with 14NO and 15NO support the assignment of a protein-bound [(μ-Cys)2Fe2(NO)4] Roussin's red ester. The [Fe4S4]2+/3+ redox couple of DNA-bound EndoIII is observable using DNA-modified gold electrochemistry, but nitrosylated EndoIII does not display observable redox activity using DNA electrochemistry on gold despite having a similar DNA-binding affinity as the native protein. However, direct electrochemistry of protein films on graphite reveals the reduction potential of native and nitrosylated EndoIII to be 127 ± 6 and -674 ± 8 mV vs NHE, respectively, corresponding to a shift of approximately -800 mV with cluster nitrosylation. Collectively, these data demonstrate that DNA-bound redox activity, and by extension DNA-mediated charge transport, is modulated by [Fe4S4] cluster nitrosylation.
Figures
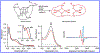


Similar articles
-
The metal core structures in the recombinant Escherichia coli transcriptional factor SoxR.Chemistry. 2012 Feb 27;18(9):2565-77. doi: 10.1002/chem.201100838. Epub 2012 Jan 20. Chemistry. 2012. PMID: 22266921
-
Nitrosylation of Nitric-Oxide-Sensing Regulatory Proteins Containing [4Fe-4S] Clusters Gives Rise to Multiple Iron-Nitrosyl Complexes.Angew Chem Int Ed Engl. 2016 Nov 14;55(47):14575-14579. doi: 10.1002/anie.201607033. Epub 2016 Oct 25. Angew Chem Int Ed Engl. 2016. PMID: 27778474 Free PMC article.
-
Reversible inactivation of E. coli endonuclease III via modification of its [4Fe-4S] cluster by nitric oxide.DNA Repair (Amst). 2003 Jul 16;2(7):809-17. doi: 10.1016/s1568-7864(03)00065-x. DNA Repair (Amst). 2003. PMID: 12826281
-
Dinitrosyl iron complexes and S-nitrosothiols are two possible forms for stabilization and transport of nitric oxide in biological systems.Biochemistry (Mosc). 1998 Jul;63(7):782-93. Biochemistry (Mosc). 1998. PMID: 9721330 Review.
-
Reactions of nitric oxide and oxygen with the regulator of fumarate and nitrate reduction, a global transcriptional regulator, during anaerobic growth of Escherichia coli.Methods Enzymol. 2008;437:191-209. doi: 10.1016/S0076-6879(07)37011-0. Methods Enzymol. 2008. PMID: 18433630 Review.
Cited by
-
The [4Fe4S] Cluster of Yeast DNA Polymerase ε Is Redox Active and Can Undergo DNA-Mediated Signaling.J Am Chem Soc. 2021 Oct 6;143(39):16147-16153. doi: 10.1021/jacs.1c07150. Epub 2021 Sep 24. J Am Chem Soc. 2021. PMID: 34559527 Free PMC article.
-
The Elusive Mononitrosylated [Fe4 S4 ] Cluster in Three Redox States.Angew Chem Int Ed Engl. 2022 Nov 21;61(47):e202213032. doi: 10.1002/anie.202213032. Epub 2022 Oct 25. Angew Chem Int Ed Engl. 2022. PMID: 36194444 Free PMC article.
-
Redox Chemistry in the Genome: Emergence of the [4Fe4S] Cofactor in Repair and Replication.Annu Rev Biochem. 2019 Jun 20;88:163-190. doi: 10.1146/annurev-biochem-013118-110644. Annu Rev Biochem. 2019. PMID: 31220976 Free PMC article. Review.
-
Cell-Penetrating Delivery of Nitric Oxide by Biocompatible Dinitrosyl Iron Complex and Its Dermato-Physiological Implications.Int J Mol Sci. 2021 Sep 18;22(18):10101. doi: 10.3390/ijms221810101. Int J Mol Sci. 2021. PMID: 34576264 Free PMC article.
-
Endogenous Conjugation of Biomimetic Dinitrosyl Iron Complex with Protein Vehicles for Oral Delivery of Nitric Oxide to Brain and Activation of Hippocampal Neurogenesis.JACS Au. 2021 Jun 7;1(7):998-1013. doi: 10.1021/jacsau.1c00160. eCollection 2021 Jul 26. JACS Au. 2021. PMID: 34467346 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

