coTRaCTE predicts co-occurring transcription factors within cell-type specific enhancers
- PMID: 30142147
- PMCID: PMC6126874
- DOI: 10.1371/journal.pcbi.1006372
coTRaCTE predicts co-occurring transcription factors within cell-type specific enhancers
Abstract
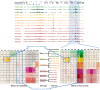
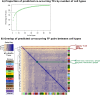
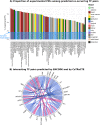
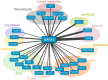
Cell-type specific gene expression is regulated by the combinatorial action of transcription factors (TFs). In this study, we predict transcription factor (TF) combinations that cooperatively bind in a cell-type specific manner. We first divide DNase hypersensitive sites into cell-type specifically open vs. ubiquitously open sites in 64 cell types to describe possible cell-type specific enhancers. Based on the pattern contrast between these two groups of sequences we develop "co-occurring TF predictor on Cell-Type specific Enhancers" (coTRaCTE) - a novel statistical method to determine regulatory TF co-occurrences. Contrasting the co-binding of TF pairs between cell-type specific and ubiquitously open chromatin guarantees the high cell-type specificity of the predictions. coTRaCTE predicts more than 2000 co-occurring TF pairs in 64 cell types. The large majority (70%) of these TF pairs is highly cell-type specific and overlaps in TF pair co-occurrence are highly consistent among related cell types. Furthermore, independently validated co-occurring and directly interacting TFs are significantly enriched in our predictions. Focusing on the regulatory network derived from the predicted co-occurring TF pairs in embryonic stem cells (ESCs) we find that it consists of three subnetworks with distinct functions: maintenance of pluripotency governed by OCT4, SOX2 and NANOG, regulation of early development governed by KLF4, STAT3, ZIC3 and ZNF148 and general functions governed by MYC, TCF3 and YY1. In summary, coTRaCTE predicts highly cell-type specific co-occurring TFs which reveal new insights into transcriptional regulatory mechanisms.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Similar articles
-
A dynamic interplay of enhancer elements regulates Klf4 expression in naïve pluripotency.Genes Dev. 2017 Sep 1;31(17):1795-1808. doi: 10.1101/gad.303321.117. Genes Dev. 2017. PMID: 28982762 Free PMC article.
-
Probing transcription factor combinatorics in different promoter classes and in enhancers.BMC Genomics. 2019 Feb 1;20(1):103. doi: 10.1186/s12864-018-5408-0. BMC Genomics. 2019. PMID: 30709337 Free PMC article.
-
Enhancer function regulated by combinations of transcription factors and cofactors.Genes Cells. 2018 Oct;23(10):808-821. doi: 10.1111/gtc.12634. Epub 2018 Aug 31. Genes Cells. 2018. PMID: 30092612 Review.
-
Predicting cell-type-specific gene expression from regions of open chromatin.Genome Res. 2012 Sep;22(9):1711-22. doi: 10.1101/gr.135129.111. Genome Res. 2012. PMID: 22955983 Free PMC article.
-
Transcription factor interactions in genomic nuclear receptor function.Epigenomics. 2011 Aug;3(4):471-85. doi: 10.2217/epi.11.66. Epigenomics. 2011. PMID: 22126206 Review.
Cited by
-
Evidence of widespread, independent sequence signature for transcription factor cobinding.Genome Res. 2021 Feb;31(2):265-278. doi: 10.1101/gr.267310.120. Epub 2020 Dec 10. Genome Res. 2021. PMID: 33303494 Free PMC article.
-
Of numbers and movement - understanding transcription factor pathogenesis by advanced microscopy.Dis Model Mech. 2020 Dec 29;13(12):dmm046516. doi: 10.1242/dmm.046516. Dis Model Mech. 2020. PMID: 33433399 Free PMC article. Review.
-
BestCRM: An Exhaustive Search for Optimal Cis-Regulatory Modules in Promoters Accelerated by the Multidimensional Hash Function.Int J Mol Sci. 2024 Feb 5;25(3):1903. doi: 10.3390/ijms25031903. Int J Mol Sci. 2024. PMID: 38339181 Free PMC article.
-
MEDEA: analysis of transcription factor binding motifs in accessible chromatin.Genome Res. 2020 May;30(5):736-748. doi: 10.1101/gr.260877.120. Epub 2020 May 18. Genome Res. 2020. PMID: 32424069 Free PMC article.
-
Integrating binding and expression data to predict transcription factors combined function.BMC Genomics. 2020 Sep 7;21(1):610. doi: 10.1186/s12864-020-06977-1. BMC Genomics. 2020. PMID: 32894066 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

