Investigation of interactions between TLR2, MyD88 and TIRAP by bioluminescence resonance energy transfer is hampered by artefacts of protein overexpression
- PMID: 30138457
- PMCID: PMC6107161
- DOI: 10.1371/journal.pone.0202408
Investigation of interactions between TLR2, MyD88 and TIRAP by bioluminescence resonance energy transfer is hampered by artefacts of protein overexpression
Abstract
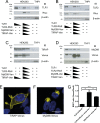
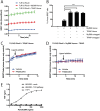
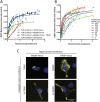
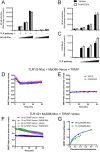
Toll like receptors (TLRs) are important pattern recognition receptors that can detect pathogen and danger associated molecular patterns to initiate an innate immune response. TLR1 and 2 heterodimerize at the plasma membrane upon binding to triacylated lipopeptides from bacterial cell walls, or to the synthetic ligand Pam3CSK4. TLR1/2 dimers interact with adaptor molecules TIRAP and MyD88 to initiate a signalling cascade that leads to activation of key transcription factors, including NF-kB. Despite TLRs being extensively studied over the last two decades, the real-time kinetics of ligand binding and receptor activation remains largely unexplored. We aimed to study the kinetics of TLR activation and recruitment of adaptors, using TLR1/2 dimer interactions with adaptors MyD88 and TIRAP. Bioluminescence resonance energy transfer (BRET) allows detection of real-time protein-protein interactions in living cells, and was applied to study adaptor recruitment to TLRs. Energy transfer showed interactions between TLR2 and TIRAP, and between TLR2 and MyD88 only in the presence of TIRAP. Quantitative BRET and confocal microscopy confirmed that TIRAP is necessary for MyD88 interaction with TLR2. Furthermore, constitutive proximity between the proteins in the absence of Pam3CSK4 stimulation was observed with BRET, and was not abrogated with lowered protein expression, changes in protein tagging strategies, or use of the brighter NanoLuc luciferase. However, co-immunoprecipitation studies did not demonstrate constitutive interaction between these proteins, suggesting that the interaction observed with BRET likely represents artefacts of protein overexpression. Thus, caution should be taken when utilizing protein overexpression in BRET studies and in investigations of the TLR pathway.
Conflict of interest statement
The authors have read the journal’s policy and have the following conflicts: KDGP is Chief Scientific Advisor of Dimerix Limited and has a shareholding in the company. KDGP receives funding from Promega, BMG Labtech and Dimerix as Partner Organisations of Australian Research Council Linkage Grant LP160100857. This does not alter the authors’ adherence to all the PLOS ONE policies on sharing data and materials. NGS, MK, LS and EME have no conflicts of interest.
Figures

Similar articles
-
The adaptor molecule TIRAP provides signalling specificity for Toll-like receptors.Nature. 2002 Nov 21;420(6913):329-33. doi: 10.1038/nature01180. Nature. 2002. PMID: 12447442
-
Molecular analysis of the binding mode of Toll/interleukin-1 receptor (TIR) domain proteins during TLR2 signaling.Mol Immunol. 2012 Oct;52(3-4):108-16. doi: 10.1016/j.molimm.2012.05.003. Epub 2012 Jun 4. Mol Immunol. 2012. PMID: 22673208
-
Essential role for TIRAP in activation of the signalling cascade shared by TLR2 and TLR4.Nature. 2002 Nov 21;420(6913):324-9. doi: 10.1038/nature01182. Nature. 2002. PMID: 12447441
-
Mal, more than a bridge to MyD88.IUBMB Life. 2013 Sep;65(9):777-86. doi: 10.1002/iub.1201. IUBMB Life. 2013. PMID: 23983209 Review.
-
Association of TLR1, TLR2, TLR4, TLR6, and TIRAP polymorphisms with disease susceptibility.Immunol Res. 2015 Jun;62(2):234-52. doi: 10.1007/s12026-015-8640-6. Immunol Res. 2015. PMID: 25784622 Review.
Cited by
-
Annexin A1 Is Required for Efficient Tumor Initiation and Cancer Stem Cell Maintenance in a Model of Human Breast Cancer.Cancers (Basel). 2021 Mar 8;13(5):1154. doi: 10.3390/cancers13051154. Cancers (Basel). 2021. PMID: 33800279 Free PMC article.
-
IL-1 Superfamily Member (IL-1A, IL-1B and IL-18) Genetic Variants Influence Susceptibility and Clinical Course of Mediterranean Spotter Fever.Biomolecules. 2022 Dec 17;12(12):1892. doi: 10.3390/biom12121892. Biomolecules. 2022. PMID: 36551320 Free PMC article.
-
NanoBRET: The Bright Future of Proximity-Based Assays.Front Bioeng Biotechnol. 2019 Mar 26;7:56. doi: 10.3389/fbioe.2019.00056. eCollection 2019. Front Bioeng Biotechnol. 2019. PMID: 30972335 Free PMC article. Review.
-
Cross-Talk between HLA Class I and TLR4 Mediates P-Selectin Surface Expression and Monocyte Capture to Human Endothelial Cells.J Immunol. 2022 Oct 1;209(7):1359-1369. doi: 10.4049/jimmunol.2200284. Epub 2022 Sep 2. J Immunol. 2022. PMID: 36165200 Free PMC article.
-
Palmitate Stimulates Expression of the von Willebrand Factor and Modulates Toll-like Receptors Level and Activity in Human Umbilical Vein Endothelial Cells (HUVECs).Int J Mol Sci. 2023 Dec 23;25(1):254. doi: 10.3390/ijms25010254. Int J Mol Sci. 2023. PMID: 38203423 Free PMC article.
References
-
- Aliprantis AO, Yang RB, Mark MR, Suggett S, Devaux B, Radolf JD, et al. Cell activation and apoptosis by bacterial lipoproteins through toll-like receptor-2. Science. 1999;285(5428):736–9. . - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

