The erlin2 T65I mutation inhibits erlin1/2 complex-mediated inositol 1,4,5-trisphosphate receptor ubiquitination and phosphatidylinositol 3-phosphate binding
- PMID: 30135210
- PMCID: PMC6177578
- DOI: 10.1074/jbc.RA118.004547
The erlin2 T65I mutation inhibits erlin1/2 complex-mediated inositol 1,4,5-trisphosphate receptor ubiquitination and phosphatidylinositol 3-phosphate binding
Abstract
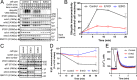
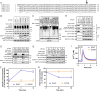
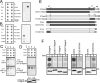
The erlin1/2 complex is a ∼2-MDa endoplasmic reticulum membrane-located ensemble of the ∼40-kDa type II membrane proteins erlin1 and erlin2. The best defined function of this complex is to mediate the ubiquitination of activated inositol 1,4,5-trisphosphate receptors (IP3Rs) and their subsequent degradation. However, it remains unclear how mutations of the erlin1/2 complex affect its cellular function and cause cellular dysfunction and diseases such as hereditary spastic paraplegia. Here, we used gene editing to ablate erlin1 or erlin2 expression to better define their individual roles in the cell and examined the functional effects of a spastic paraplegia-linked mutation to erlin2 (threonine to isoleucine at position 65; T65I). Our results revealed that erlin2 is the dominant player in mediating the interaction between the erlin1/2 complex and IP3Rs and that the T65I mutation dramatically inhibits this interaction and the ability of the erlin1/2 complex to promote IP3R ubiquitination and degradation. Remarkably, we also discovered that the erlin1/2 complex specifically binds to phosphatidylinositol 3-phosphate, that erlin2 binds this phospholipid much more strongly than does erlin1, that the binding is inhibited by T65I mutation of erlin2, and that multiple determinants within the erlin2 polypeptide comprise the phosphatidylinositol 3-phosphate-binding site. Overall, these results indicate that erlin2 is the primary mediator of the cellular roles of the erlin1/2 complex and that disease-linked mutations of erlin2 can affect both IP3R processing and lipid binding.
Keywords: endoplasmic reticulum (ER); endoplasmic-reticulum-associated protein degradation (ERAD); erlin1; erlin2; inositol trisphosphate receptor (InsP3R); mutation; phosphatidylinositol phosphatase; phospholipid signaling; spastic paraplegia; ubiquitylation (ubiquitination).
© 2018 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures
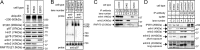
Similar articles
-
Binding of the erlin1/2 complex to the third intralumenal loop of IP3R1 triggers its ubiquitin-proteasomal degradation.J Biol Chem. 2022 Jun;298(6):102026. doi: 10.1016/j.jbc.2022.102026. Epub 2022 May 11. J Biol Chem. 2022. PMID: 35568199 Free PMC article.
-
RNF170 protein, an endoplasmic reticulum membrane ubiquitin ligase, mediates inositol 1,4,5-trisphosphate receptor ubiquitination and degradation.J Biol Chem. 2011 Jul 8;286(27):24426-33. doi: 10.1074/jbc.M111.251983. Epub 2011 May 24. J Biol Chem. 2011. PMID: 21610068 Free PMC article.
-
Bi-allelic variants in RNF170 are associated with hereditary spastic paraplegia.Nat Commun. 2019 Oct 21;10(1):4790. doi: 10.1038/s41467-019-12620-9. Nat Commun. 2019. PMID: 31636353 Free PMC article.
-
Chapter 4 - Inositol 1,4,5-Trisphosphate Receptor Ubiquitination.Prog Mol Biol Transl Sci. 2016;141:141-59. doi: 10.1016/bs.pmbts.2016.02.004. Epub 2016 Mar 30. Prog Mol Biol Transl Sci. 2016. PMID: 27378757 Review.
-
A novel homozygous mutation in ERLIN1 gene causing spastic paraplegia 62 and literature review.Eur J Med Genet. 2022 Nov;65(11):104608. doi: 10.1016/j.ejmg.2022.104608. Epub 2022 Sep 12. Eur J Med Genet. 2022. PMID: 36100157 Review.
Cited by
-
Binding of the erlin1/2 complex to the third intralumenal loop of IP3R1 triggers its ubiquitin-proteasomal degradation.J Biol Chem. 2022 Jun;298(6):102026. doi: 10.1016/j.jbc.2022.102026. Epub 2022 May 11. J Biol Chem. 2022. PMID: 35568199 Free PMC article.
-
Transcriptome and Literature Mining Highlight the Differential Expression of ERLIN1 in Immune Cells during Sepsis.Biology (Basel). 2021 Aug 5;10(8):755. doi: 10.3390/biology10080755. Biology (Basel). 2021. PMID: 34439987 Free PMC article.
-
The quantification of zebrafish ocular-associated proteins provides hints for sex-biased visual impairments and perception.Heliyon. 2024 Jun 13;10(12):e33057. doi: 10.1016/j.heliyon.2024.e33057. eCollection 2024 Jun 30. Heliyon. 2024. PMID: 38994070 Free PMC article.
-
The emerging link between IP3 receptor turnover and Hereditary Spastic Paraplegia.Cell Calcium. 2020 Mar;86:102142. doi: 10.1016/j.ceca.2019.102142. Epub 2019 Dec 18. Cell Calcium. 2020. PMID: 31874412 Free PMC article.
-
Role of ERLINs in the Control of Cell Fate through Lipid Rafts.Cells. 2021 Sep 13;10(9):2408. doi: 10.3390/cells10092408. Cells. 2021. PMID: 34572057 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

