Stabilization of p27Kip1/CDKN1B by UBCH7/UBE2L3 catalyzed ubiquitinylation: a new paradigm in cell-cycle control
- PMID: 30113882
- PMCID: PMC6355086
- DOI: 10.1096/fj.201800960R
Stabilization of p27Kip1/CDKN1B by UBCH7/UBE2L3 catalyzed ubiquitinylation: a new paradigm in cell-cycle control
Abstract
Ubiquitinylation drives many cellular processes by targeting proteins for proteasomal degradation. Ubiquitin conjugation enzymes promote ubiquitinylation and, thus, degradation of protein substrates. Ubiquitinylation is a well-known posttranslational modification controlling cell-cycle transitions and levels or/and activation levels of ubiquitin-conjugating enzymes change during development and cell cycle. Progression through the cell cycle is tightly controlled by CDK inhibitors such as p27Kip1. Here we show that, in contrast to promoting its degradation, the ubiquitin-conjugating enzyme UBCH7/UBE2L3 specifically protects p27Kip1 from degradation. Overexpression of UBCH7/UBE2L3 stabilizes p27Kip1 and delays the G1-to-S transition, while depletion of UBCH7/UBE2L3 increases turnover of p27Kip1. Levels of p21Cip1/Waf1, p57Kip2, cyclin A and cyclin E, all of which are also involved in regulating the G1/S transition are not affected by UBCH7/UBE2L3 depletion. The effect of UBCH7/UBE2L3 on p27Kip1 is not due to alteration of the levels of any of the ubiquitin ligases known to ubiquitinylate p27Kip1. Rather, UBCH7/UBE2L3 catalyzes the conjugation of heterotypic ubiquitin chains on p27Kip1 that are proteolytically incompetent. These data reveal new controls and concepts about the ubiquitin proteasome system in which a ubiquitin-conjugating enzyme selectively inhibits and may even protect, rather than promote degradation of a crucial cell-cycle regulatory molecule.-Whitcomb, E. A., Tsai, Y. C., Basappa, J., Liu, K., Le Feuvre, A. K., Weissman, A. M., Taylor, A. Stabilization of p27Kip1/CDKN1B by UBCH7/UBE2L3 catalyzed ubiquitinylation: a new paradigm in cell-cycle control.
Keywords: CDKI; cell division; proteasome; ubiquitin-conjugating enzyme.
Conflict of interest statement
Funding for this work was provided by U.S. National Institutes of Health (NIH) National Eye Institute Grants RO1 EY 13250, RO1 EY21212, and RO1 EY26979, U.S. Department of Agriculture contract 1950-510000-060-03A (to A.T.), and the NIH Center for Cancer Research–National Cancer Institute Intramural Research Program (to Y.C.T. and A.M.W.). The authors thank E. Bejarano-Fernandez (Tufts University) for critical review and W. Yang (Brandeis University, Waltham, MA, USA) for expert technical assistance. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. The authors declare no conflicts of interest.
Figures

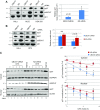


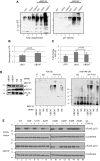
Similar articles
-
The E3 ligase SMURF1 stabilizes p27 via UbcH7 catalyzed K29-linked ubiquitin chains to promote cell migration SMURF1-UbcH7 K29 ubiquitination of p27 and cell migration.J Biol Chem. 2024 Mar;300(3):105693. doi: 10.1016/j.jbc.2024.105693. Epub 2024 Feb 1. J Biol Chem. 2024. PMID: 38301893 Free PMC article.
-
Ubiquitylation and proteasomal degradation of the p21(Cip1), p27(Kip1) and p57(Kip2) CDK inhibitors.Cell Cycle. 2010 Jun 15;9(12):2342-52. doi: 10.4161/cc.9.12.11988. Epub 2010 Jun 15. Cell Cycle. 2010. PMID: 20519948 Free PMC article. Review.
-
Damage-specific DNA binding protein 1 (DDB1) is involved in ubiquitin-mediated proteolysis of p27Kip1 in response to UV irradiation.Biochimie. 2011 May;93(5):867-75. doi: 10.1016/j.biochi.2010.12.017. Epub 2011 Jan 13. Biochimie. 2011. PMID: 21237244
-
Kip1 degradation via the ubiquitin-proteasome pathway.Leukemia. 1997 Apr;11 Suppl 3:363-6. Leukemia. 1997. PMID: 9209391
-
Mechanism and Disease Association With a Ubiquitin Conjugating E2 Enzyme: UBE2L3.Front Immunol. 2022 Feb 21;13:793610. doi: 10.3389/fimmu.2022.793610. eCollection 2022. Front Immunol. 2022. PMID: 35265070 Free PMC article. Review.
Cited by
-
Proteostasis in aging-associated ocular disease.Mol Aspects Med. 2022 Dec;88:101157. doi: 10.1016/j.mam.2022.101157. Epub 2022 Nov 29. Mol Aspects Med. 2022. PMID: 36459837 Free PMC article. Review.
-
KAT2A/E2F1 Promotes Cell Proliferation and Migration via Upregulating the Expression of UBE2C in Pan-Cancer.Genes (Basel). 2022 Oct 8;13(10):1817. doi: 10.3390/genes13101817. Genes (Basel). 2022. PMID: 36292703 Free PMC article.
-
Integrated Analysis of Transcriptome and Metabolome Reveals Distinct Responses of Pelteobagrus fulvidraco against Aeromonas veronii Infection at Invaded and Recovering Stage.Int J Mol Sci. 2022 Sep 4;23(17):10121. doi: 10.3390/ijms231710121. Int J Mol Sci. 2022. PMID: 36077519 Free PMC article.
-
A structurally conserved site in AUP1 binds the E2 enzyme UBE2G2 and is essential for ER-associated degradation.PLoS Biol. 2021 Dec 8;19(12):e3001474. doi: 10.1371/journal.pbio.3001474. eCollection 2021 Dec. PLoS Biol. 2021. PMID: 34879065 Free PMC article.
-
From the Evasion of Degradation to Ubiquitin-Dependent Protein Stabilization.Cells. 2021 Sep 9;10(9):2374. doi: 10.3390/cells10092374. Cells. 2021. PMID: 34572023 Free PMC article. Review.
References
-
- Liu Q., Shang F., Zhang X., Li W., Taylor A. (2006) Expression of K6W-ubiquitin inhibits proliferation of human lens epithelial cells. Mol. Vis. 12, 931–936 - PubMed
-
- Liu K., Lyu L., Chin D., Gao J., Sun X., Shang F., Caceres A., Chang M. L., Rowan S., Peng J., Mathias R., Kasahara H., Jiang S., Taylor A. (2015) Altered ubiquitin causes perturbed calcium homeostasis, hyperactivation of calpain, dysregulated differentiation, and cataract. Proc. Natl. Acad. Sci. USA 112, 1071–1076; erratum: 112, E817 - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

